Chapter 3 Data Wrangling
So far in our journey,
we’ve seen how to look at data saved in data frames
using the dplyr::glimpse() or View() functions
in Chapter 1,
and how to create data visualizations using the ggplot2 package
in Chapter 2.
In particular we studied what we term the “five named graphs” (5NG):
- scatterplots via
geom_point() - linegraphs via
geom_line() - boxplots via
geom_boxplot() - histograms via
geom_histogram() - barplots via
geom_bar()orgeom_col()
We created these visualizations using the grammar of graphics,
which maps variables in a data frame
to the aesthetic attributes of one of the 5 geometric objects.
We can also control other aesthetic attributes of the geometric objects
such as the size and color as seen in the Gapminder data example
in Figure 2.1.
Recall however that for two of our visualizations,
we first needed to transform/modify existing data frames a little.
For example, recall the scatterplot in Figure 2.2 of departure
and arrival delays only for Alaska Airlines flights.
In order to create this visualization,
we first needed to pare down the df_flights data frame
to an df_alaska_flights data frame
consisting of only carrier == "AS" flights.
Thus, df_alaska_flights will have fewer rows than df_flights.
We did this using the filter() function:
In this chapter, we’ll extend this example
and we’ll introduce a series of functions from the dplyr package
for data wrangling that will allow you to take a data frame
and “wrangle” it (transform it) to suit your needs.
Such functions include:
filter()a data frame’s existing rows to only pick out a subset of them. For example, wrangle/transformdf_flightstodf_alaska_flights.select()a data frame’s existing columns to only pick out a subset of them.summarize()one or more of its columns/variables with a summary statistic. Examples of summary statistics include the median and interquartile range of temperatures as we saw in Section 2.7 on boxplots.group_by()rows. This function is often used together with another function to achieve a “per-group” effect. For example, we can combinegroup_by()withsummarize()to report summary statistics for each group separately. Say you want to calculate the average departure delaydep_delayfor each of the threeoriginairports indf_alaska_flights. You can do so by first assigning all rows into three groups withgroup_by(), followed by applyingsummarize(). You will get three separate average departure delays, one computed for each of the threeoriginairports. Without applyinggroup_by()first, you would only get a single overall average departure delaydep_delayfor all threeoriginairports combined,mutate()an existing columns/variables to create new ones. For example, convert hourly temperature recordings from degrees Fahrenheit to degrees Celsius.arrange()rows in a specific order. For example, sort the rows ofdf_weatherin ascending or descending order oftemp.join()a data frame with another data frame by matching along a “key” variable. In other words, merge these two data frames together.
Notice how we used computer_code font to describe the actions
we want to take on our data frames.
This is because the dplyr package for data wrangling
has intuitively verb-named functions that are easy to remember.
There is a further benefit to learning to use the dplyr package for data wrangling: its similarity to the database querying language SQL (pronounced “sequel” or spelled out as “S,” “Q,” “L”). SQL (which stands for “Structured Query Language”) is used to manage large databases quickly and efficiently and is widely used by many institutions with a lot of data. While SQL is a topic left for a book or a course on database management, keep in mind that once you learn dplyr, you can learn SQL easily. We’ll talk more about their similarities in Subsection 3.8.1.4.
Needed packages
The following code gives us access to the demo data, as well as the to some tools for us to interact with the data.
# Install xfun so that I can use xfun::pkg_load2
if (!requireNamespace('xfun')) install.packages('xfun')
xf <- loadNamespace('xfun')
cran_primary <- c(
"dplyr",
"ggplot2",
"nycflights13"
)
if (length(cran_primary) != 0) xf$pkg_load2(cran_primary)
import::from(magrittr, "%>%")
gg <- import::from(ggplot2, .all=TRUE, .into={new.env()})
dp <- import::from(dplyr, .all=TRUE, .into={new.env()})
import::from(nycflights13,
df_airlines = airlines,
df_airports = airports,
df_flights = flights,
df_planes = planes,
df_weather = weather
)
# Create the Celsius column by ourselves as it does not come with df_weather
df_weather$temp_c = (df_weather$temp - 32) * (5 / 9)3.1 The pipe operator: %>%
Before we start data wrangling,
let’s first introduce a nifty tool:
the pipe operator %>%.
We have used it in various places over the past two weeks,
but a formal introduction is in order.
As ubiqutous as it is, the pipe operator %>% is not one of base R functions.
Instead, it is part of package magrittr.
Therefore, we import it before using it in our code:
The pipe operator allows us to combine multiple operations in R
into a single sequential chain of actions.
Let’s start with a hypothetical example.
Say you would like to perform a hypothetical sequence of operations
on a hypothetical data frame x using hypothetical functions
f(), g(), and h():
- Take
xthen - Use
xas an input to a functionf()then - Use the output of
f(x)as an input to a functiong()then - Use the output of
g(f(x))as an input to a functionh()
One way to achieve this sequence of operations is by using nesting parentheses as follows:
This code isn’t so hard to read since we are applying only three functions:
f(), then g(), then h() and each of the functions is short in its name.
Further, each of these functions also only has one argument.
However, you can imagine that this will get progressively harder to read
as the number of functions applied in your sequence increases
and the arguments in each function increase as well.
This is where the pipe operator %>% comes in handy.
%>% takes the output of one function
and then “pipes” it to be the input of the next function.
Furthermore, a helpful trick is to read %>% as “then” or “and then.”
For example, you can obtain the same output
as the hypothetical sequence of functions as follows:
You would read this sequence as:
- Take
xthen - Use this output as the input to the next function
f()then - Use this output as the input to the next function
g()then - Use this output as the input to the next function
h()
So while both approaches achieve the same goal,
the latter is much more human-readable
because you can clearly read the sequence of operations line-by-line.
But what are the hypothetical x, f(), g(), and h()?
Throughout this chapter on data wrangling:
- The starting value
xwill be a data frame. For example, thedf_flightsdata frame we explored in Section 1.4. - The sequence of functions, here
f(),g(), andh(), will mostly be a sequence of any number of the data wrangling verb-named functions we listed in the introduction to this chapter. For example, thedplyr::filter(carrier == "AS")function and argument specified we previewed earlier. - The result will be the transformed/modified data frame that you want.
In our example, we’ll save the result in a new data frame
by using the
<-assignment operator as in:df_alaska_flights <-.
df_alaska_flights <- df_flights %>%
dp$filter(carrier == "AS")
# reminder: earlier, we chose `dp` as a shorthand for `dplyr`,Much like when adding layers to a ggplot() using the + sign,
you form a single chain of data wrangling operations
by combining verb-named functions into a single sequence
using the pipe operator %>%.
Furthermore, much like how the + sign has to come
at the end of lines when constructing plots,
the pipe operator %>% has to come at the end of lines as well.
Keep in mind, there are many more advanced data wrangling functions than just the ones listed in the introduction to this chapter; you’ll see some examples of these in Section 3.8. However, just with these seven verb-named functions you’ll be able to perform a broad array of data wrangling tasks for the rest of this course.
3.2 filter rows
The filter in a coffee machine,
the filter in a vaccum,
or the filter in a water jug.
What do they have in common?
They all stop unwanted elements and only let the “good stuff” through.
Similarly, a filter function lets you
selectively keep observations that interest you
and drop the rest.
The filter() function in dplyr packages works
much like the “Filter” option in Microsoft Excel;
it allows you to specify criteria about the values of a variable
in your dataset and then filters out only the rows that match that criteria.
We begin by focusing only on flights from New York City to Portland, Oregon.
The dest destination code (or airport code) for Portland, Oregon is "PDX".
Run the following and look at the results
to ensure that only flights heading to Portland are chosen here:
df_portland_flights <- df_flights %>%
dp$filter(dest == "PDX")
# use the `unique()` function to list all the unique values in a column
unique(df_portland_flights$dest)
# Alternatively, you can open the data frame and eyeball it
# View(df_portland_flights)Note the order of the code.
Start from the right side of the assignment arrow <-.
First take the df_flights data frame,
then filter() the data frame
so that only those where the dest equals "PDX" are included.
We test for equality using the double equal sign ==,
not a single equal sign =.
This is a convention across many programming languages.
If you are new to coding,
you’ll probably forget to use the double equal sign == a few times
before you get the hang of it.
You can use other operators
beyond just the == operator that tests for equality:
>corresponds to “greater than”<corresponds to “less than”>=corresponds to “greater than or equal to”<=corresponds to “less than or equal to”!=corresponds to “not equal to.” The!is used in many programming languages to indicate “not.”
Furthermore, you can combine multiple criteria with these two special operators:
|corresponds to “or” (shift + backward slash\on most US keyboards)&corresponds to “and”
To see another example,
let’s filter df_flights for all rows that
- departed from JFK
- and were heading to Burlington, Vermont ("BTV")
or Seattle, Washington ("SEA")
- and departed in the months of October, November, or December.
Run the following:
btv_sea_flights_fall <- df_flights %>%
dp$filter(origin == "JFK" & (dest == "BTV" | dest == "SEA") & month >= 10)
View(btv_sea_flights_fall)Note that even though colloquially speaking
one might say “all flights leaving Burlington, Vermont
and Seattle, Washington,”
in terms of computer operations,
we really mean “all flights leaving Burlington, Vermont
or leaving Seattle, Washington.”
For a given row in the data, dest can be "BTV", or "SEA",
or something else, but not both "BTV" and "SEA" at the same time.
Furthermore, note the careful use of parentheses
around dest == "BTV" | dest == "SEA".
For filter(), We can skip the use of &
and just separate our conditions with a comma.
The previous code will return the identical output btv_sea_flights_fall
as the following code:
btv_sea_flights_fall <- df_flights %>%
dp$filter(origin == "JFK", (dest == "BTV" | dest == "SEA"), month >= 10)
str(btv_sea_flights_fall)Let’s present another example that uses the !
“not” operator to pick rows that don’t match a criteria.
As mentioned earlier, the ! can be read as “not.”
Here we are filtering rows corresponding to flights that
didn’t go to Burlington, VT or Seattle, WA.
Again, note the careful use of parentheses
around the (dest == "BTV" | dest == "SEA").
If we didn’t use parentheses as follows:
We would be returning all flights not headed to "BTV"
or those headed to "SEA",
which is an entirely different resulting data frame.
Now say we have a larger number of airports we want to filter for,
say "SEA", "SFO", "PDX", "BTV", and "BDL".
We could continue to use the | (or) operator:
many_airports <- df_flights %>%
dp$filter(dest == "SEA" | dest == "SFO" | dest == "PDX" |
dest == "BTV" | dest == "BDL")but as we progressively include more airports,
this will get unwieldy to write.
A slightly shorter approach uses the %in% operator
along with the c() function.
Recall from Subsection 1.2.1
that the c() function “combines” or “concatenates” values
into a single vector of values.
many_airports <- df_flights %>%
dp$filter(dest %in% c("SEA", "SFO", "PDX", "BTV", "BDL"))
str(many_airports)What this code is doing is filtering df_flights for all flights
where dest is in the vector of airports
c("BTV", "SEA", "PDX", "SFO", "BDL").
Both outputs of many_airports are the same,
but as you can see the latter takes much less energy to code.
The %in% operator is useful for looking for matches
commonly in one vector/variable compared to another.
As a final note, we recommend that
filter() should often be among the first verbs you consider
applying to your data.
This narrows down your dataset to only those rows you care about,
before you further transform the data with other operations.
Learning check
(LC3.1) Use a single equal sign on purpose where a double equal sign is needed in the following code, and observe the error message you will get.
# Run on your own and observe what happens
df_portland_flights <- df_flights %>%
dp$filter(dest = "PDX")(LC3.2)
What’s another way of using the “not” operator !
to filter only the rows that are not going to Burlington, VT
nor Seattle, WA in the df_flights data frame?
Test this out using the previous code.
3.3 select variables
We have seen how filter lets us pick rows from a data frame.
Next, we will learn to use select for selecting columns from a data frame.
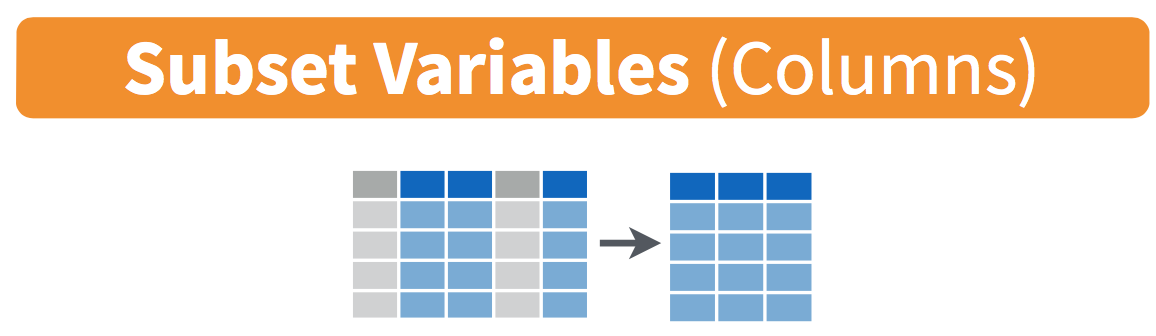
Figure 3.1: Diagram of select() columns.
The df_flights data frame in the nycflights13 package
contains 19 different variables.
You can identify the names of these 19 variables
by running the glimpse() function from the dplyr package:
However, say you only need two of these 19 variables,
carrier and flight.
You can select() these two variables:
This function makes it easier to explore large datasets since it allows us to limit the scope to only those variables we care most about.
Let’s say instead you want to drop, or de-select, certain variables.
For example, consider the variable year in the df_flights data frame.
This variable isn’t quite a “variable”
because it is always 2013 and hence doesn’t change.
Say you want to remove this variable from the data frame.
We can deselect year by using the - sign:
Another way of selecting columns/variables is by specifying a range of columns:
This will select() all columns between month and day,
as well as between arr_time and sched_arr_time, and drop the rest.
The select() function can also be used to reorder columns
when used with the everything() helper function.
For example, suppose we want the hour, minute, and time_hour variables
to appear immediately after the year, month, and day variables,
while not discarding the rest of the variables.
In the following code, everything() will pick up all remaining variables:
flights_reorder <- df_flights %>%
dp$select(year, month, day, hour, minute, time_hour, dp$everything())
dp$glimpse(flights_reorder)Lastly, the helper functions starts_with(), ends_with(), and contains()
can be used to select variables/columns that match those conditions.
As examples,
df_flights %>% dp$select(dp$starts_with("a"))
df_flights %>% dp$select(dp$ends_with("delay"))
df_flights %>% dp$select(dp$contains("time"))Learning check
(LC3.3)
What are some ways to select all three of the dest, air_time,
and distance variables from df_flights?
Give the code showing how to do this in at least three different ways.
(LC3.4)
How could one use starts_with(), ends_with(), and contains()
to select columns from the df_flights data frame?
Provide three different examples in total:
one for starts_with(), one for ends_with(), and one for contains().
(LC3.5)
Why might we want to use the select function on a data frame?
3.4 summarize variables
The next common task when working with data frames is to compute summary statistics. Common examples of summary statistics include mean (also known as average), median, variance, to name a few. Each summary statistic is a single numerical value that aggregates over a large number of values. Take the mean of a sample for example. When we calculate the average height of 50 adults, we gain some insight about the heights of those 50 people from the single numerical value — the average height: How tall is a typical person in this group of 50 people?
Other examples of summary statistics that might not immediately come to mind include the sum, the smallest value also called the minimum, the largest value also called the maximum, and the standard deviation. See Appendix A.1 for a glossary of such summary statistics.
Let’s calculate two summary statistics of the temp_c temperature variable
in the df_weather data frame:
the mean and standard deviation
(recall from Section 1.4
that we have seen the df_weather data frame).
To compute these summary statistics,
we need the mean() and sd() summary functions in R.
A summary function in R takes in many values and returns a single value,
as illustrated in Figure 3.2.
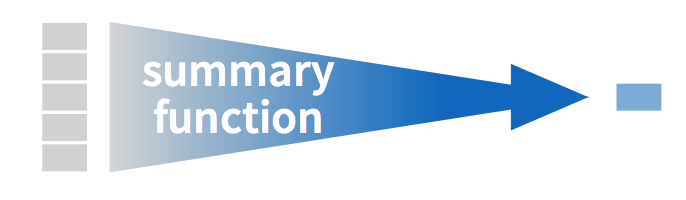
Figure 3.2: Diagram illustrating a summary function in R.
We’ll use the mean() and sd() summary functions
within the summarize() function
from the dplyr package.
Note you can also use the British English spelling of summarise().
We’ll save the results in a new data frame called summary_temp
that will have two columns/variables: the mean and the std_dev:
# A tibble: 1 x 2
mean std_dev
<dbl> <dbl>
1 NA NAWhy are the values returned NA?
As we saw in Subsection 2.3.1
when creating the scatterplot of departure and arrival delays
for df_alaska_flights,
NA indicates “not available” or “not applicable”
and it is how R encodes missing values.
If a value in a particular cell does not exist, NA is stored instead.
Values can be missing for many reasons.
Perhaps the observation was collected by someone
who forgot to enter it?
Perhaps the observation was not collected at all
because it was too difficult to do so?
Perhaps there was an erroneous value that someone entered
and has been replaced with NA?
You’ll often encounter issues with missing values when working with real data.
Going back to our summary_temp output,
by default any time you try to calculate a summary statistic of a variable
that has one or more NA missing values in R, NA is returned.
To work around this fact, you can set the na.rm argument to TRUE,
where rm is short for “remove”;
this will ignore any NA missing values
and only return the summary value for all non-missing values.
The code that follows computes the mean and standard deviation
of all non-missing values of temp_c:
summary_temp <- df_weather %>%
dp$summarize(mean = mean(temp_c, na.rm = TRUE),
std_dev = sd(temp_c, na.rm = TRUE))
summary_temp# A tibble: 1 x 2
mean std_dev
<dbl> <dbl>
1 12.9 9.88Notice how the na.rm = TRUE are used
as arguments to each of summary statistic functions
such as the mean() and sd() ,
not to the summarize() function.
One needs to be cautious whenever ignoring missing values
as we’ve just done.
In the upcoming Learning checks questions,
we’ll consider the possible ramifications of blindly sweeping rows
with missing values “under the rug.”
This is in fact why the na.rm argument to any summary statistic function
in R is set to FALSE by default.
When using R, we as researchers will be forced to make a conscious decision
about any missing values that may exist.
In other words, R does not ignore rows with missing values by default.
R will alert you to the presence of missing data.
You should be mindful of this missingness
and its potential causes throughout your analysis.
What are other summary functions we can use inside the summarize() verb
to compute summary statistics?
As seen in the diagram in Figure 3.2,
you can use any function in R that takes many values
and returns just one. Here are just a few:
mean(): the averagesd(): the standard deviation, which is a measure of spreadmin()andmax(): the minimum and maximum values, respectivelyIQR(): interquartile rangesum(): the total amount when adding multiple numbersn(): a count of the number of rows in each group. This particular summary function will make more sense whengroup_by()is covered in Section 3.5.
Note that all but the last function list above are part of base R,
and therefore can be used without reference to their packages.
n() is a function from the package dplyr,
and thus needs to be explicitly referenced, as in dplyr::n()
or dp$n() for short.
Learning check
(LC3.6) Say a doctor is studying the effect of smoking on lung cancer for a large number of patients who have records measured at five-year intervals. She notices that a large number of patients have missing data points because the patient has died, so she chooses to ignore these patients in her analysis. What is wrong with this doctor’s approach?
(LC3.7)
Modify the earlier summarize() function code
that creates the summary_temp data frame
to also use the n() summary function:
summarize(... , count = dp$n()).
What does the returned value correspond to?
(LC3.8) Why doesn’t the following code work?
df_weather %>%
dp$summarize(mean = mean(temp_c, na.rm = TRUE)) %>%
dp$summarize(std_dev = sd(temp_c, na.rm = TRUE))Hint: To diagnose, run the code in two steps instead of all at once,
and look at the result in each step.
First, run
Then add back the second statement:
df_weather %>%
dp$summarize(mean = mean(temp_c, na.rm = TRUE)) %>%
dp$summarize(std_dev = sd(temp_c, na.rm = TRUE))(LC3.9)
What does the value calculated by mean(df_weather$temp_c, na.rm=TRUE) represent?
3.5 group_by rows
Say instead of a single mean temperature for the whole year,
you would like 12 mean temperatures,
one for each of the 12 months separately.
Put differently, we would like to compute the mean temperature split by month.
We can do this by “grouping” temperature observations
by the values of another variable,
in this case by the 12 values of the variable month.
Run the following code:
summary_monthly_temp <- df_weather %>%
dp$group_by(month) %>%
dp$summarize(mean = mean(temp_c, na.rm = TRUE),
std_dev = sd(temp_c, na.rm = TRUE))
summary_monthly_temp# A tibble: 12 x 3
month mean std_dev
* <int> <dbl> <dbl>
1 1 2.02 5.68
2 2 1.26 3.88
3 3 4.38 3.47
4 4 11.0 4.88
5 5 16.6 5.38
6 6 22.3 4.19
7 7 26.7 3.96
8 8 23.6 2.88
9 9 19.7 4.70
10 10 15.6 4.91
11 11 7.22 5.80
12 12 3.58 5.55This code is almost identical to the previous code that created summary_temp,
with an extra dp$group_by(month) added before the summarize().
Grouping the df_weather dataset by month
and then applying the summarize() functions
yields a data frame that displays the mean and standard deviation temperature
split by the 12 months of the year.
It is important to note that the
group_by() function does not result in a transformed data frame
the same way filter() or summarize() does.
Instead, it changes the meta-data, or data about the data,
specifically the grouping structure.
It is only after we apply the summarize() function, for example, that
a new data frame gets created.
Let’s consider the diamonds data frame
included in the ggplot2 package. Run this code:
# A tibble: 53,940 x 10
carat cut color clarity depth table price x y z
<dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
4 0.290 Premium I VS2 62.4 58 334 4.2 4.23 2.63
5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
7 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47
8 0.26 Very Good H SI1 61.9 55 337 4.07 4.11 2.53
9 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49
10 0.23 Very Good H VS1 59.4 61 338 4 4.05 2.39
# … with 53,930 more rowsObserve that the first line of the output reads
# A tibble: 53,940 x 10.
This is an example of meta-data,
in this case the number of observations/rows and variables/columns
in df_diamonds.
The actual data itself are the subsequent table of values.
Now let’s pipe the df_diamonds data frame into group_by(cut):
# A tibble: 53,940 x 10
# Groups: cut [5]
carat cut color clarity depth table price x y z
<dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
4 0.290 Premium I VS2 62.4 58 334 4.2 4.23 2.63
5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
7 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47
8 0.26 Very Good H SI1 61.9 55 337 4.07 4.11 2.53
9 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49
10 0.23 Very Good H VS1 59.4 61 338 4 4.05 2.39
# … with 53,930 more rowsObserve that now there is additional meta-data:
# Groups: cut [5] indicating that
the grouping structure meta-data has been set
based on the 5 possible levels of the categorical variable cut:
"Fair", "Good", "Very Good", "Premium", and "Ideal".
On the other hand, observe that the data has not changed:
it is still a table of 53,940
\(\times\) 10 values.
Only by combining a group_by() with another data wrangling operation,
in this case summarize(), will the data actually be transformed.
# A tibble: 5 x 2
cut avg_price
* <ord> <dbl>
1 Fair 4359.
2 Good 3929.
3 Very Good 3982.
4 Premium 4584.
5 Ideal 3458.Let’s now revisit the n() counting summary function
we briefly introduced previously.
Recall that the n() function counts rows.
This is opposed to the sum() summary function
that returns the sum of a numerical variable.
For example, suppose we’d like to count
how many flights departed each of the three airports in New York City:
# A tibble: 3 x 2
origin count
* <chr> <int>
1 EWR 120835
2 JFK 111279
3 LGA 104662We see that Newark ("EWR") had the most flights departing in 2013
followed by "JFK" and lastly by LaGuardia ("LGA").
Note there is a subtle but important difference between sum() and n();
while sum() returns the sum of a numerical variable,
n() returns a count of the number of rows/observations.
Learning check
(LC3.10)
Run the following code and compare its output to by_origin.
What do you think is the purpose of the function count?
Verify your guess by reading its help page
https://dplyr.tidyverse.org/reference/count.html
3.5.1 Grouping by more than one variable
You are not limited to grouping by one variable.
Say you want to know the number of flights
leaving each of the three New York City airports for each month.
We can also group by a second variable month using group_by(origin, month):
by_origin_monthly <- df_flights %>%
dp$group_by(origin, month) %>%
dp$summarize(count = dp$n())
by_origin_monthly# A tibble: 36 x 3
# Groups: origin [3]
origin month count
<chr> <int> <int>
1 EWR 1 9893
2 EWR 2 9107
3 EWR 3 10420
4 EWR 4 10531
5 EWR 5 10592
6 EWR 6 10175
7 EWR 7 10475
8 EWR 8 10359
9 EWR 9 9550
10 EWR 10 10104
# … with 26 more rowsWhen you run this code on your own, you might receive a message like this:
`summarise()` has grouped output by 'origin'.
You can override using the `.groups` argument.You can safely ignore this message for now. If you are curious about what this message means, check out this stackoverflow post.
Observe that there are 36 rows
in by_origin_monthly
because there are 12 months for 3 airports (EWR, JFK, and LGA).
Why do we group_by(origin, month)
and not group_by(origin) and then group_by(month)?
Let’s investigate:
by_origin_monthly_incorrect <- df_flights %>%
dp$group_by(origin) %>%
dp$group_by(month) %>%
dp$summarize(count = dp$n())
by_origin_monthly_incorrect# A tibble: 12 x 2
month count
* <int> <int>
1 1 27004
2 2 24951
3 3 28834
4 4 28330
5 5 28796
6 6 28243
7 7 29425
8 8 29327
9 9 27574
10 10 28889
11 11 27268
12 12 28135What happened here is that the second group_by(month)
overwrote the grouping structure meta-data of the earlier group_by(origin),
so that in the end we are only grouping by month.
The lesson here is if you want to group_by() two or more variables,
you should include all the variables at the same time in the same group_by()
adding a comma between the variable names.
Learning check
(LC3.11)
Recall from Chapter 2 when we looked at temperatures by months in NYC.
What does the standard deviation column
in the summary_monthly_temp data frame (see Section 3.5) tell us
about temperatures in NYC throughout the year?
(LC3.12) What code would be required to get the mean and standard deviation temperature for each day in 2013 for NYC?
(LC3.13)
Recreate by_monthly_origin,
but instead of grouping via group_by(origin, month),
group variables in a different order group_by(month, origin).
What differs in the resulting dataset?
(LC3.14)
How could we identify how many flights left each of the three airports
for each carrier?
3.6 mutate existing variables

Figure 3.3: Diagram of mutate() columns.
Another common transformation of data is to create/compute new variables
based on existing ones.
For example, in Section 2.4 we converted temp
which was in Fahrenheit, to temp_c in Celsius.
The formula to convert temperatures from °F to °C is
\[ \text{temp in C} = \frac{\text{temp in F} - 32}{1.8} \]
We can apply this formula to the temp variable
using the mutate() function from the dplyr package,
which takes existing variables and mutates them to create new ones.
Good to know
Notice the df_weather <- part.
Without it, the rest of code reads
df_weather %>% dp$mutate(temp_c = (temp - 32) / 1.8),
which in English loosely translates to:
take the data frame df_weather, transform the column temp
with mutate and save the resulting column in temp_c.
So why do we need the df_weather <- part?
If you are not already familiar with in-place versus not-in-place modification,
I encourage you to go through the following experiment
and deduce the answer from its result.
- Step 1: convert
temptotemp_c, but drop thedf_weather <-part:
# start from scratch and import the data frame again
import::from(nycflights13, df_weather = weather)
# convert Fahrenheit to Celsius
df_weather %>% dp$mutate(temp_c = (temp - 32) / 1.8)- step 2: examine the columns of
df_weather. Specifically, istemp_camong them?
- step 3: confirm that
temp_ctruly is not present
- step 4: re-run step 1 but this time with
df_weather <-part:
- step 5: verify that
temp_cis now present
What can you deduce from these results?
In the code above, we mutate() the df_weather data frame
by creating a new variable temp_c = (temp - 32) / 1.8
and then overwrite the original df_weather data frame
with all the original columns plus the newly created temp_c column.
Why did we overwrite the data frame df_weather,
instead of assigning the result to a new data frame like weather_new?
As a rough rule of thumb,
as long as you are not losing original information that you might need later,
it’s acceptable to overwrite existing data frames with updated ones,
as we did here.
On the other hand, why did we NOT overwrite the variable temp,
but instead created a new variable called temp_c?
Because if we did this,
we would have erased the original information contained
in temp of temperatures in Fahrenheit that may still be valuable to us.
Let’s now compute monthly average temperatures
in both °F and °C using the group_by() and summarize() code
we saw in Section 3.5:
summary_monthly_temp <- df_weather %>%
dp$group_by(month) %>%
dp$summarize(mean_temp_in_F = mean(temp, na.rm = TRUE),
mean_temp_c = mean(temp_c, na.rm = TRUE))
summary_monthly_temp# A tibble: 12 x 3
month mean_temp_in_F mean_temp_c
* <int> <dbl> <dbl>
1 1 35.6 2.02
2 2 34.3 1.26
3 3 39.9 4.38
4 4 51.7 11.0
5 5 61.8 16.6
6 6 72.2 22.3
7 7 80.1 26.7
8 8 74.5 23.6
9 9 67.4 19.7
10 10 60.1 15.6
11 11 45.0 7.22
12 12 38.4 3.58Let’s consider another example.
Passengers are often frustrated when their flight departs late,
but aren’t as annoyed if, in the end,
pilots can make up some time during the flight.
This is known in the airline industry as gain,
and we will create this variable using the mutate() function:
Let’s take a look at only the dep_delay, arr_delay,
and the resulting gain variables for the first 5 rows
in our updated df_flights data frame in Table 3.1.
| dep_delay | arr_delay | gain |
|---|---|---|
| 2 | 11 | -9 |
| 4 | 20 | -16 |
| 2 | 33 | -31 |
| -1 | -18 | 17 |
| -6 | -25 | 19 |
The flight in the first row departed 2 minutes late but arrived 11 minutes late,
so its “gained time in the air” is a loss of 9 minutes,
hence its gain is \(2 - 11 = -9\).
On the other hand, the flight in the fourth row
departed a minute early (dep_delay of -1)
but arrived 18 minutes early (arr_delay of -18),
so its “gained time in the air” is \(-1 - (-18) = -1 + 18 = 17\) minutes,
hence its gain is +17.
Let’s look at some summary statistics of the gain variable
by considering multiple summary functions at once
in the same summarize() code:
gain_summary <- df_flights %>%
dp$summarize(
min = min(gain, na.rm = TRUE),
q1 = quantile(gain, 0.25, na.rm = TRUE),
median = quantile(gain, 0.5, na.rm = TRUE),
q3 = quantile(gain, 0.75, na.rm = TRUE),
max = max(gain, na.rm = TRUE),
mean = mean(gain, na.rm = TRUE),
sd = sd(gain, na.rm = TRUE),
missing = sum(is.na(gain))
)
gain_summary# A tibble: 1 x 8
min q1 median q3 max mean sd missing
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int>
1 -196 -3 7 17 109 5.66 18.0 9430We see for example that the average gain is +5 minutes,
while the largest is +109 minutes!
However, this code would take some time to type out in practice.
We’ll see later on in Subsection 11.1
that there is a much more succinct way to compute
a variety of common summary statistics:
using the skim() function from the skimr package.
Recall from Section 2.5
that since gain is a continuous variable,
we can visualize its distribution using a histogram.
gg$ggplot(data = df_flights,
mapping = gg$aes(x = gain)) +
gg$geom_histogram(color = "white", bins = 20)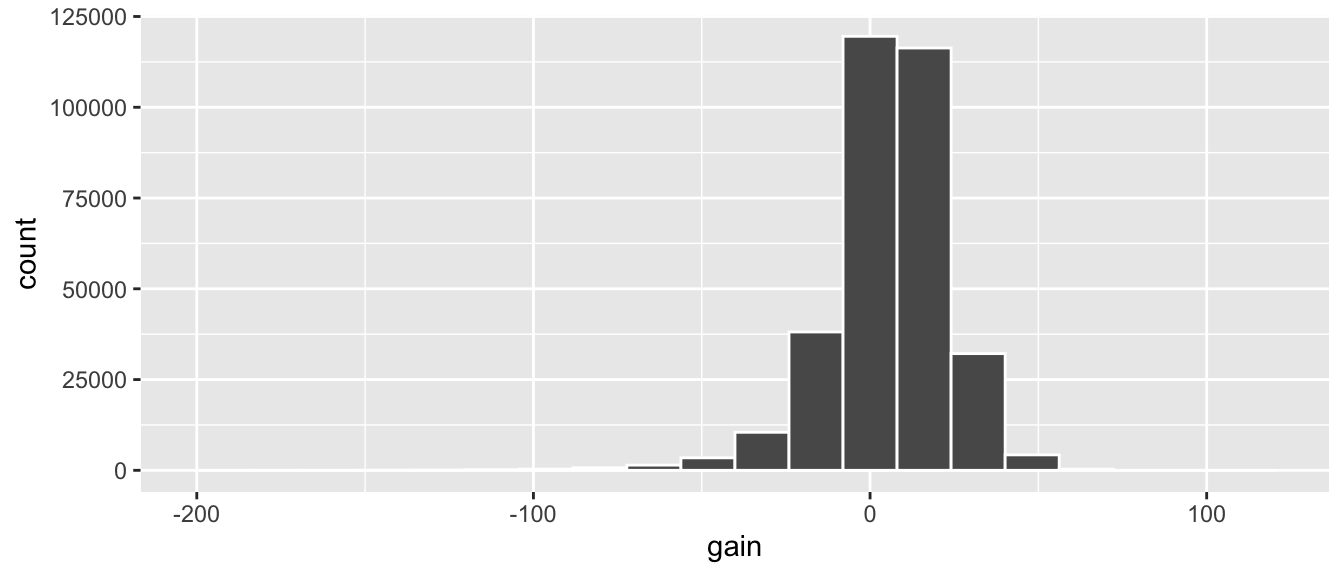
Figure 3.4: Histogram of gain variable.
The resulting histogram in Figure 3.4
provides a different perspective on the gain variable
than the summary statistics we computed earlier.
For example, note that most values of gain are right around 0.
To close out our discussion on the mutate() function,
note that we can create multiple new variables at once
in the same mutate() code.
Furthermore, within the same mutate() code
we can refer to new variables we just created.
As an example, consider the mutate() code
Hadley Wickham and Garrett Grolemund
show in Chapter 5 of R for Data Science (Grolemund and Wickham 2017):
df_flights <- df_flights %>%
dp$mutate(
gain = dep_delay - arr_delay,
hours = air_time / 60,
gain_per_hour = gain / hours
)Learning check
(LC3.15)
What do positive values of the gain variable in df_flights correspond to?
What about negative values? And what about a zero value?
(LC3.16)
Could we create the dep_delay and arr_delay columns
by simply subtracting dep_time from sched_dep_time
and similarly for arrivals?
Try the code out and explain any differences
between the result and what actually appears in df_flights.
(LC3.17)
What can we say about the distribution of gain?
Describe it in a few sentences using the plot
and the gain_summary data frame values.
3.7 arrange and sort rows
One of the most commonly performed data wrangling tasks
is to sort a data frame’s rows in the alphanumeric order
of one of the variables.
The dplyr package’s arrange() function allows us
to sort/reorder a data frame’s rows
according to the values of the specified variable.
Suppose we are interested in determining the most frequent destination airports for all domestic flights departing from New York City in 2013:
# A tibble: 105 x 2
dest num_flights
* <chr> <int>
1 ABQ 254
2 ACK 265
3 ALB 439
4 ANC 8
5 ATL 17215
6 AUS 2439
7 AVL 275
8 BDL 443
9 BGR 375
10 BHM 297
# … with 95 more rowsObserve that by default the rows of the resulting freq_dest data frame
are sorted in alphabetical order of destination.
Say instead we would like to see the same data,
but sorted from the most to the least number of flights (num_flights) instead:
# A tibble: 105 x 2
dest num_flights
<chr> <int>
1 LEX 1
2 LGA 1
3 ANC 8
4 SBN 10
5 HDN 15
6 MTJ 15
7 EYW 17
8 PSP 19
9 JAC 25
10 BZN 36
# … with 95 more rowsThis is, however, the opposite of what we want.
The rows are sorted with the least frequent destination airports displayed first.
This is because arrange() always returns rows
sorted in ascending order by default.
To switch the ordering to be in “descending” order instead,
we use the desc() function as so:
# A tibble: 105 x 2
dest num_flights
<chr> <int>
1 ORD 17283
2 ATL 17215
3 LAX 16174
4 BOS 15508
5 MCO 14082
6 CLT 14064
7 SFO 13331
8 FLL 12055
9 MIA 11728
10 DCA 9705
# … with 95 more rows3.8 Other verbs
Here are some other useful data wrangling verbs:
join()two different datasets.rename()variables/columns to have new names.slice()the highest/lowestnrows of a variable
3.8.1 join data frames
A common data transformation task is “joining” or “merging”
two different datasets.
For example, you just conducted a study on working memory
in which you collected response time
from participants on several different tasks.
At the end of the study,
participants also filled out a demographic questionnaire.
The response time and demographic information were stored
in two separate spreadsheet-like files, as is often the case.
To test hypotheses that involve both response time and
participants’ age, for example,
we need to join the two aforementioned data frames.
In this section, we will learn another function, join,
using datasets from the nycflights13 package as an example.
In the df_flights data frame,
the variable carrier lists the carrier code for the different flights.
While the corresponding airline names for "UA" and "AA"
might be somewhat easy to guess (United and American Airlines),
what airlines have codes "VX", "HA", and "B6"?
This information is provided in a separate data frame airlines
from the package nycflights13. Let’s import it and save it as df_airlines.
We see that in df_airlines, carrier is the carrier code,
while name is the full name of the airline company.
Using this table, we can see that "VX", "HA", and "B6"
correspond to Virgin America, Hawaiian Airlines, and JetBlue, respectively.
However, wouldn’t it be nice to have all this information
in df_flights instead of having to look it up in a separate data frame
such as df_airlines?
We can do this by “joining” the df_flights and df_airlines data frames.
Note that the values in the variable carrier in the df_flights data frame
match the values in the variable carrier in the df_airlines data frame.
In this case, we can use the variable carrier
as a key variable
to match the rows of the two data frames.
Key variables are almost always identification variables
that uniquely identify the observational units
as we saw in Subsection 1.4.3.
This ensures that rows in both data frames are appropriately matched
during the join.
Hadley and Garrett (Grolemund and Wickham 2017) created the diagram shown
in Figure 3.5 to help us understand
how the different data frames in the nycflights13 package
are linked by various key variables:
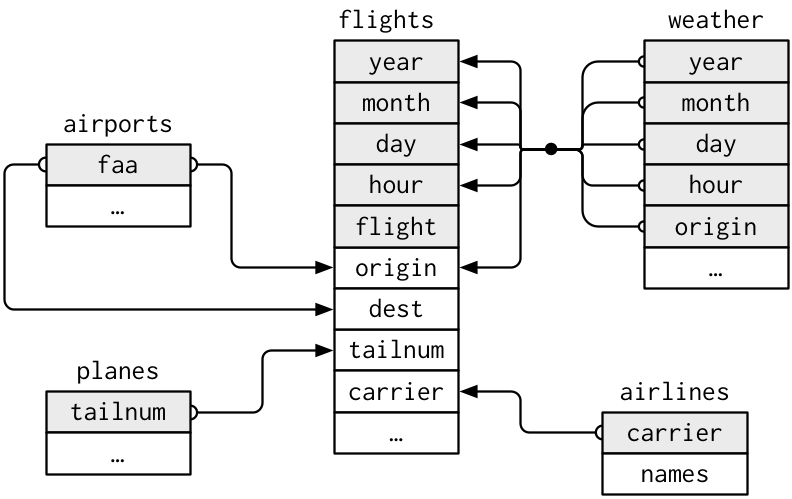
Figure 3.5: Data relationships in nycflights13 from R for Data Science.
3.8.1.1 Matching “key” variable names
In both the df_flights and df_airlines data frames,
the key variable we want to join/merge/match the rows by
has the same name: carrier.
Let’s use the inner_join() function
to join the two data frames,
where the rows will be matched by the variable carrier,
and then compare the resulting data frames:
flights_joined <- df_flights %>%
dp$inner_join(df_airlines, by = "carrier")
str(df_flights)
str(flights_joined)Observe that the df_flights and flights_joined data frames
are identical except that flights_joined has an additional variable name.
The values of name correspond to the airline companies’ names
as indicated in the df_airlines data frame.
A visual representation of the inner_join() is shown
in Figure 3.6 (Grolemund and Wickham 2017).
There are other types of joins available
(such as left_join(), right_join(), outer_join(), and anti_join()),
but the inner_join() will solve nearly all of the problems
you’ll encounter in this book.
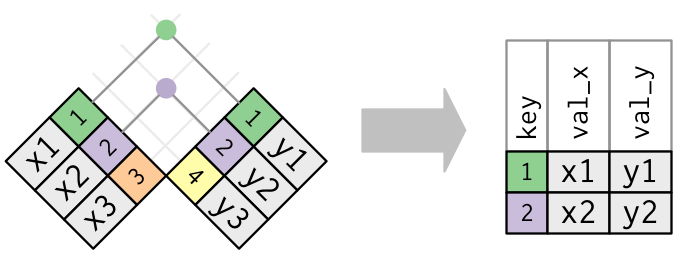
Figure 3.6: Diagram of inner join from R for Data Science.
3.8.1.2 Different “key” variable names
Say instead you are interested in the destinations of all domestic flights
departing NYC in 2013, and you ask yourself questions like:
“What cities are these airports in?”
or “Is "ORD" Orlando?” or “Where is "FLL"?”
The airports data frame from package nycflights13
contains the airport codes for each airport:
However, if you look at both the df_airports and df_flights data frames,
you’ll find that the airport codes are in variables that have different names.
In df_airports the airport code is in faa,
whereas in df_flights the airport codes are in origin and dest.
This fact is further highlighted in the visual representation
of the relationships between these data frames in Figure 3.5.
In order to join these two data frames by airport code,
our inner_join() operation will use the by = c("dest" = "faa")
argument with modified code syntax
allowing us to join two data frames where the key variable has a different name:
flights_with_airport_names <- df_flights %>%
dp$inner_join(df_airports, by = c("dest" = "faa"))
str(flights_with_airport_names)Let’s construct the chain of pipe operators %>%
that computes the number of flights from NYC to each destination,
but also includes information about each destination airport:
named_dests <- df_flights %>%
dp$group_by(dest) %>%
dp$summarize(num_flights = dp$n()) %>%
dp$arrange(dp$desc(num_flights)) %>%
dp$inner_join(df_airports, by = c("dest" = "faa")) %>%
dp$rename(airport_name = name)
named_dests# A tibble: 101 x 9
dest num_flights airport_name lat lon alt tz dst tzone
<chr> <int> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 ORD 17283 Chicago Ohare Intl 42.0 -87.9 668 -6 A America…
2 ATL 17215 Hartsfield Jackson… 33.6 -84.4 1026 -5 A America…
3 LAX 16174 Los Angeles Intl 33.9 -118. 126 -8 A America…
4 BOS 15508 General Edward Law… 42.4 -71.0 19 -5 A America…
5 MCO 14082 Orlando Intl 28.4 -81.3 96 -5 A America…
6 CLT 14064 Charlotte Douglas … 35.2 -80.9 748 -5 A America…
7 SFO 13331 San Francisco Intl 37.6 -122. 13 -8 A America…
8 FLL 12055 Fort Lauderdale Ho… 26.1 -80.2 9 -5 A America…
9 MIA 11728 Miami Intl 25.8 -80.3 8 -5 A America…
10 DCA 9705 Ronald Reagan Wash… 38.9 -77.0 15 -5 A America…
# … with 91 more rowsIn case you didn’t know,
"ORD" is the airport code of Chicago O’Hare airport
and "FLL" is the main airport in Fort Lauderdale, Florida,
which can be seen in the airport_name variable.
3.8.1.3 Multiple “key” variables
Say instead we want to join two data frames by multiple key variables.
For example, in Figure 3.5,
we see that in order to join the flights and weather data frames,
we need more than one key variable:
year, month, day, hour, and origin.
This is because the combination of these 5 variables
act to uniquely identify each observational unit
in the weather data frame:
hourly weather recordings at each of the 3 NYC airports.
We achieve this by specifying a vector of key variables
to join by using the c() function.
Recall from Subsection 1.2.1
that c() is short for “combine” or “concatenate.”
flights_weather_joined <- df_flights %>%
dp$inner_join(df_weather, by = c("year", "month", "day", "hour", "origin"))
str(flights_weather_joined)Learning check
(LC3.18)
Looking at Figure 3.5,
when joining flights and weather
(or, in other words, matching the hourly weather values with each flight),
why do we need to join by all of year, month, day, hour, and origin,
and not just hour?
(LC3.19) What surprises you about the top 10 destinations from NYC in 2013?
3.8.1.4 Normal forms
The data frames included in the nycflights13 package are in a form
that minimizes redundancy of data.
For example, the df_flights data frame only saves the carrier code
of the airline company;
it does not include the actual name of the airline.
For example, the first row of df_flights has carrier equal to UA,
but it does not include the airline name of “United Air Lines Inc.”
The names of the airline companies are included in the name variable
of the df_airlines data frame.
In order to have the airline company name included in df_flights,
we could join these two data frames as follows:
We are capable of performing this join
because each of the data frames have keys in common
to relate one to another:
the carrier variable in both the df_flights and df_airlines data frames.
The key variable(s) that we base our joins on
are often identification variables as we mentioned previously.
This is an important property of what’s known as normal forms of data. The process of decomposing data frames into less redundant tables without losing information is called normalization. More information is available on Wikipedia.
Both dplyr and SQL
we mentioned in the introduction of this chapter use such normal forms.
Given that they share such commonalities,
once you learn either of these two tools,
you can learn the other very easily.
Learning check
(LC3.20) What are some advantages of data in normal forms? What are some disadvantages?
3.8.2 rename variables
Another useful function is rename(),
which as you may have guessed changes the name of variables.
Suppose we want to only focus on dep_time and arr_time
and change dep_time and arr_time to be departure_time and arrival_time
in the flights_time data frame:
flights_time_new <- df_flights %>%
dp$select(dep_time, arr_time) %>%
dp$rename(departure_time = dep_time, arrival_time = arr_time)
dp$glimpse(flights_time_new)Note that in this case we used a single = sign within the rename().
For example, departure_time = dep_time renames the dep_time variable
to have the new name departure_time.
This is because we are not testing for equality
like we would using ==.
Instead we want to assign a new variable departure_time
to have the same values as dep_time
and then delete the variable dep_time.
Note that new dplyr users often forget
that the new variable name comes before the equal sign,
as in “new = old.”
3.8.3 slice select rows with highest/lowest values of a variable
We can also slice the first/last n rows ordered by a certain variable.
Take named_dests from Subsection 3.8.1.2 as an example.
We can use the slice_max function
to return a data frame of the top 10 destination airports.
Observe that we set order_by = num_flights
and the number of values to return to n = 10
to indicate that we want the rows corresponding
to the top 10 values of num_flights.
What about the lowest values of a variable?
As you may have guessed, there is also a slice_min()
which does exactly that.
Note that slice_max() and slice_min() may return more rows than requested
in the presence of ties.
You can use with_ties = FALSE to suppress.
Learning check
(LC3.21) Create a new data frame that shows the top 5 airports with the largest arrival delays from NYC in 2013.
(LC3.22)
Find out what function slice_head() does by reading its help page
https://dplyr.tidyverse.org/reference/slice.html.
What about slice_sample()?
3.9 Conclusion
3.9.1 Summary table
Let’s recap our data wrangling verbs in Table 3.2.
Using these verbs and the pipe %>% operator from Section 3.1,
you’ll be able to write easily legible code
to perform almost all the data wrangling
and data transformation necessary for the rest of this course.
| Verb | Data wrangling operation |
|---|---|
filter()
|
Stipulate which rows to ratain |
select()
|
Stipulate which columns to retain or drop |
summarize()
|
Summarize many values to one using a summary statistic function like mean(), median(), etc.
|
group_by()
|
Add grouping structure to rows in data frame. Note this does not change values in data frame, rather only the meta-data |
mutate()
|
Create new variables by mutating existing ones |
arrange()
|
Arrange rows of a data variable in ascending (default) or descending order
|
rename()
|
change column names |
slice()
|
retain the highest/lowest n values of a variable |
inner_join()
|
Join/merge two data frames, matching rows by a key variable |
Learning check
(LC3.23) Let’s now put your newly acquired data wrangling skills to the test!
An airline industry measure of a passenger airline’s capacity is the available seat miles, which is equal to the number of seats available multiplied by the number of miles flown summed over all flights.
For example, let’s consider the scenario in Figure 3.7. Since the airplane has 4 seats and it travels 200 miles, the available seat miles are \(4 \times 200 = 800\).
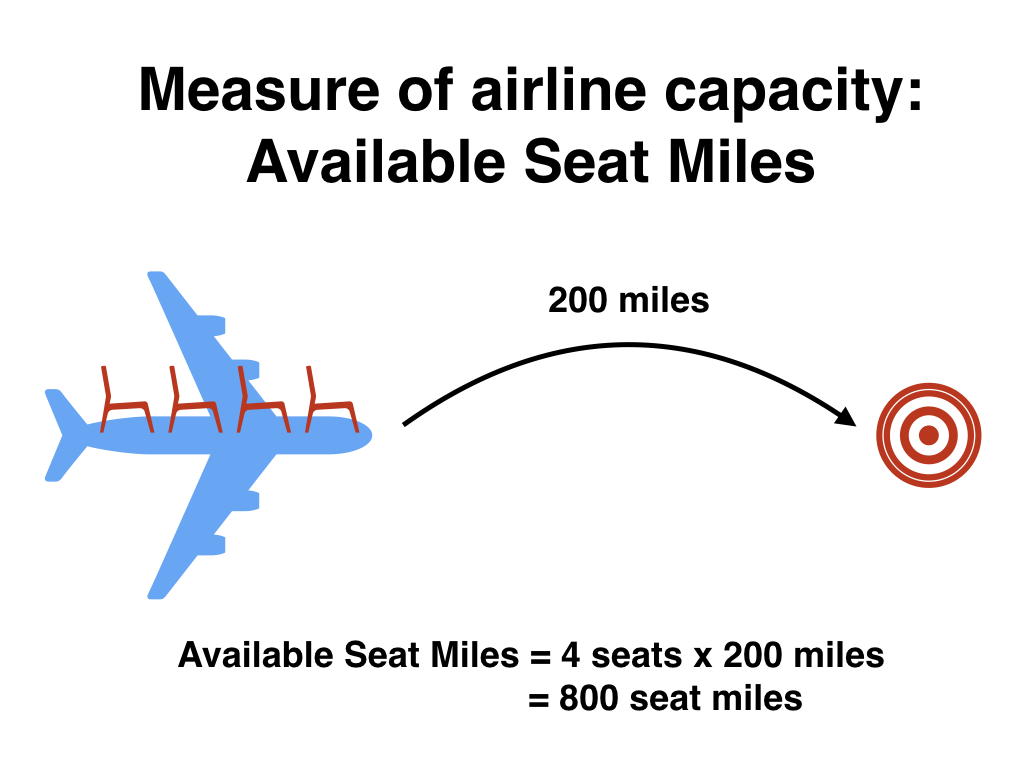
Figure 3.7: Example of available seat miles for one flight.
Extending this idea, let’s say an airline had 2 flights using a plane with 10 seats that flew 500 miles and 3 flights using a plane with 20 seats that flew 1000 miles, the available seat miles would be \(2 \times 10 \times 500 + 3 \times 20 \times 1000 = 70,000\) seat miles.
Using the datasets included in the nycflights13 package,
compute the available seat miles for each airline sorted in descending order.
After completing all the necessary data wrangling steps,
the resulting data frame should have 16 rows (one for each airline)
and 2 columns (airline name and available seat miles).
Here are some hints:
- Crucial: Unless you are very confident in what you are doing,
it is worthwhile not starting to code right away.
Rather, first sketch out on paper all the necessary data wrangling steps
not using exact code,
but rather high-level pseudocode that is informal
yet detailed enough to articulate what you are doing.
This way you won’t confuse what you are trying to do (the algorithm)
with how you are going to do it (writing
dplyrcode). - Take a close look at all the datasets using the
View()function:df_flights,df_weather,df_planes,df_airports, anddf_airlinesto identify which variables are necessary to compute available seat miles. - Figure 3.5 showing how the various datasets can be joined will also be useful.
- Consider the data wrangling verbs in Table 3.2 as your toolbox!
3.9.2 Additional resources
On top of the data wrangling verbs and examples we presented in this section,
if you’d like to see more examples of using the dplyr package
for data wrangling,
check out Chapter 5
of R for Data Science (Grolemund and Wickham 2017).
3.9.3 What’s to come?
So far in this book, we’ve explored, visualized, and wrangled data saved in data frames. These data frames were saved in a spreadsheet-like format: in a rectangular shape with a certain number of rows corresponding to observations and a certain number of columns corresponding to variables describing these observations.
We’ll see in the upcoming Chapter 4 that there are actually two ways to represent data in spreadsheet-type rectangular format: (1) “wide” format and (2) “tall/narrow” format. The tall/narrow format is also known as “tidy” format in R user circles. While the distinction between “tidy” and non-“tidy” formatted data is subtle, it has immense implications for our data science work. This is because almost all the packages used in this book, including the ggplot2 package for data visualization and the dplyr package for data wrangling, all assume that all data frames are in “tidy” format.
Furthermore, up until now we’ve only explored, visualized, and wrangled data saved within R packages. But what if you want to analyze data that you have saved in a Microsoft Excel, a Google Sheets, or a “Comma-Separated Values” (CSV) file? In Section 4.1, we’ll show you how to import this data into R using the readr package.