Chapter 2 Data Visualization
We begin the development of your data science toolbox with data visualization.
By visualizing data,
we gain valuable insights we couldn’t initially obtain
from just looking at the raw data values.
We’ll use the ggplot2 package,
as it provides an easy way to customize your plots.
ggplot2 is rooted in the data visualization theory
known as the grammar of graphics (Wilkinson 2005),
developed by Leland Wilkinson.
At their most basic, graphics/plots/charts (we use these terms interchangeably in this book) provide a nice way to explore the patterns in data, such as the presence of outliers, distributions of individual variables, and relationships between groups of variables. Graphics are designed to emphasize the findings and insights you want your audience to understand. This does, however, require a balancing act. On the one hand, you want to highlight as many interesting findings as possible. On the other hand, you don’t want to include so much information that it overwhelms your audience.
As we will see, plots also help us to identify patterns and outliers in our data. We’ll see that a common extension of these ideas is to compare the distribution of one continuous variable, such as what are the center and spread of the values, as we go across the levels of a different categorical variable.
Needed packages
We will be playing with a few demo datasets throughout this chapter. The following code gives us access to the data, as well as the to some tools so we can interact with the data.
# Install xfun so that I can use xfun::pkg_load2
if (!requireNamespace('xfun')) install.packages('xfun')
xf <- loadNamespace('xfun')
cran_primary <- c(
"dplyr",
"gapminder",
"ggplot2",
"nycflights13",
"tibble"
)
if (length(cran_primary) != 0) xf$pkg_load2(cran_primary)
gg <- import::from(ggplot2, .all=TRUE, .into={new.env()})
dp <- import::from(dplyr, .all=TRUE, .into={new.env()})
import::from(magrittr, "%>%")2.1 The grammar of graphics
We start with a discussion of a theoretical framework for data visualization
known as “the grammar of graphics.”
This framework serves as the foundation for the
ggplot2 package which we’ll use extensively in this chapter.
Think of how we construct and form sentences in English
by combining different elements,
like nouns, verbs, articles, subjects, objects, etc.
We can’t just combine these elements in any arbitrary order;
we must do so following a set of rules known as a linguistic grammar.
Similarly to a linguistic grammar,
“the grammar of graphics” defines a set of rules
for constructing statistical graphics
by combining different types of layers.
This grammar was created by Leland Wilkinson (Wilkinson 2005)
and has been implemented in a variety of data visualization software platforms
like R, but also Plotly
and Tableau.
2.1.1 Components of the grammar
In short, the grammar tells us that:
A statistical graphic is a
mappingofdatavariables toaesthetic attributes ofgeometric objects.
Specifically, we can break a graphic into the following three essential components:
data: the dataset containing the variables of interest.geom: the geometric object in question. This refers to the type of object we can observe in a plot. For example: points, lines, and bars.aes: aesthetic attributes of the geometric object. For example, x/y position, color, shape, and size.
Aesthetic attributes are mapped to variables in the dataset.
You might be wondering why we wrote the terms data, geom, and aes
in a computer code type font.
We’ll see very shortly that we’ll specify the elements of the grammar in R
using these terms.
However, let’s first break down the grammar with an example.
2.1.2 Gapminder data
In February 2006, a Swedish physician and data advocate named Hans Rosling gave a TED talk titled “The best stats you’ve ever seen” where he presented global economic, health, and development data from the website gapminder.org. For example, for data on 142 countries in 2007, let’s consider only a few countries in Table 2.1 as a peek into the data.
| Country | Continent | Life Expectancy | Population | GDP per Capita |
|---|---|---|---|---|
| Afghanistan | Asia | 43.83 | 31889923 | 974.58 |
| Albania | Europe | 76.42 | 3600523 | 5937.03 |
| Algeria | Africa | 72.30 | 33333216 | 6223.37 |
Each row in this table corresponds to a country in 2007. For each row, we have 5 columns:
Country: Name of country.
Continent: Which of the five continents the country is part of. Note that “Americas” includes countries in both North and South America and that Antarctica is excluded.
Life Expectancy: Life expectancy in years.
Population: Number of people living in the country.
GDP per Capita: Gross domestic product (in US dollars).
Now consider Figure 2.1, which plots this for all 142 of the data’s countries.
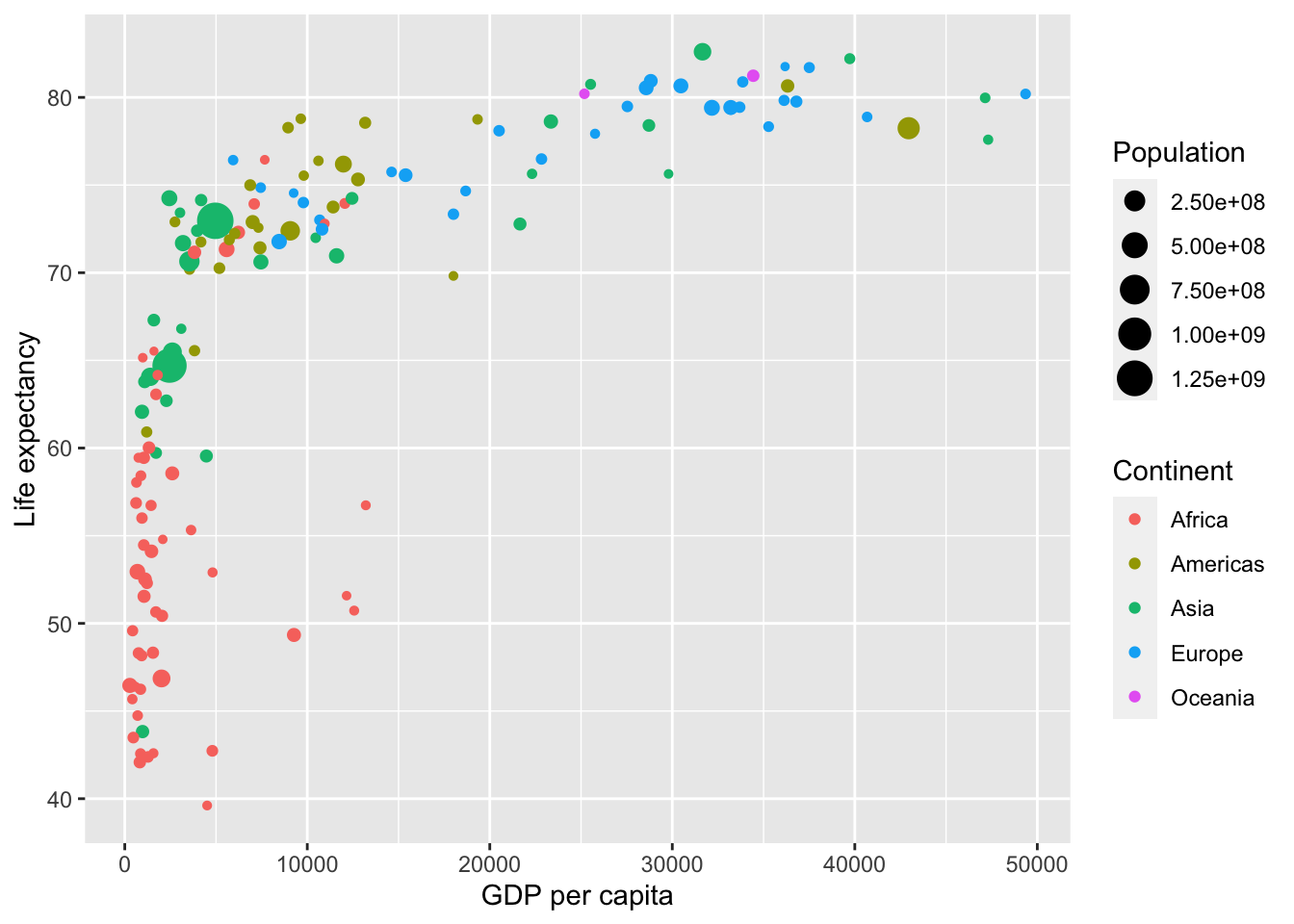
Figure 2.1: Life expectancy over GDP per capita in 2007.
Let’s view this plot through the grammar of graphics:
The
datavariable GDP per Capita gets mapped to thex-positionaesthetic of the points.The
datavariable Life Expectancy gets mapped to they-positionaesthetic of the points.The
datavariable Population gets mapped to thesizeaesthetic of the points.The
datavariable Continent gets mapped to thecoloraesthetic of the points.
We’ll see shortly that data corresponds to the particular data frame
where our data is saved and
that “data variables” correspond to particular columns in the data frame.
Furthermore, the type of geometric object considered
in this plot are points.
That being said, while in this example we are considering points,
graphics are not limited to just points.
We can also use lines, bars, and other geometric objects.
Let’s summarize the three essential components of the grammar in Table 2.2.
| data variable | aes | geom |
|---|---|---|
| GDP per Capita | x | point |
| Life Expectancy | y | point |
| Population | size | point |
| Continent | color | point |
2.1.3 Other components
There are other components of the grammar of graphics we can control as well.
As you start to delve deeper into the grammar of graphics,
you’ll start to encounter these topics more frequently.
In this book, we’ll keep things simple
and only work with these two additional components:
faceting breaks up a plot into several plots split by the values of another variable (Section 2.6)positionadjustments for barplots (Section 2.8)
Other more complex components like scales and coordinate systems
are left for a more advanced text such as
R for Data Science
(Grolemund and Wickham 2017).
Generally speaking, the grammar of graphics
allows for a high degree of customization of plots
and also a consistent framework for easily updating and modifying them.
2.1.4 ggplot2 package
In this book, we will use the ggplot2 package for data visualization,
which is an implementation of the grammar of graphics for R
(Wickham et al. 2020).
As we noted earlier, a lot of the previous section was written
in a computer code type font.
This is because the various components of the grammar of graphics
are specified in the ggplot() function
included in the ggplot2 package.
For the purposes of this book,
we’ll always provide the ggplot() function with the following arguments
(i.e., inputs) at a minimum:
- The data frame where the variables exist: the
dataargument. - The mapping of the variables to aesthetic attributes:
the
mappingargument which specifies theaesthetic attributes involved.
After we’ve specified these components,
we then add layers to the plot using the + sign.
The most essential layer to add to a plot is the layer
that specifies which type of geometric object we want the plot to involve:
points, lines, bars, and others.
Other layers we can add to a plot include the plot title,
axes labels, visual themes for the plots,
and facets (which we’ll see in Section 2.6).
Let’s now put the theory of the grammar of graphics into practice.
2.2 Five named graphs - the 5NG
In order to keep things simple in this book, we will only focus on five different types of graphics, each with a commonly given name. We term these “five named graphs” or in abbreviated form, the 5NG:
- scatterplots
- linegraphs
- histograms
- boxplots
- barplots
We’ll also present some variations of these plots, but with this basic repertoire of five graphics in your toolbox, you can visualize a wide array of different variable types. Note that certain plots are only appropriate for categorical variables, while others are only appropriate for continuous variables.
2.3 5NG#1: Scatterplots
The simplest of the 5NG are scatterplots,
also called bivariate plots.
They allow you to visualize the relationship between two continuous variables.
While you may already be familiar with scatterplots,
let’s view them through the lens of the grammar of graphics
we presented in Section 2.1.
Specifically, we will visualize the relationship
between the following two continuous variables in the flights data frame
included in the nycflights13 package:
dep_delay: departure delay on the horizontal “x” axis andarr_delay: arrival delay on the vertical “y” axis
As we did before in Chapter 1.4,
let’s first import flights and save it as df_flights.
Next, let’s pare down the data from all 336,776 flights that left NYC in 2013, to only the 714 Alaska Airlines flights that left NYC in 2013. We do this so our scatterplot will involve a manageable 714 points, and not an overwhelmingly large number like 336,776.
To achieve this,
we’ll take the newly imported df_flights data frame,
filter the rows so that only the 714 rows
corresponding to Alaska Airlines flights are kept,
and save this in a new data frame called df_alaska_flights
using the <- assignment operator:
This code above uses the dplyr package for data wrangling to achieve our goal:
it takes the df_flights data frame
and filters it to only return the rows
where carrier is equal to "AS",
Alaska Airlines’ carrier code.
Recall from Section 1.2 that
testing for equality is specified with ==
and not =.
This filtering action is one type of data wrangling,
a topic we will discuss in detail in Chapter 3.
Now try it yourself
and explore the resulting data frame by running View(df_alaska_flights).
You’ll see that it has 714 rows,
consisting of only 714 Alaska Airlines flights.
Now you should be convinced that the code above
achieved what it was supposed to.
2.3.1 Scatterplots via geom_point
Let’s now go over the code that will create the desired scatterplot, while keeping in mind the grammar of graphics framework we introduced in Section 2.1. Let’s take a look at the code and break it down piece-by-piece.
gg$ggplot(data = df_alaska_flights,
mapping = gg$aes(x = dep_delay, y = arr_delay)) +
gg$geom_point()Within the ggplot() function,
we specify two of the components of the grammar of graphics
as arguments (i.e., inputs):
- The
dataas thedf_alaska_flightsdata frame viadata = df_alaska_flights. - The
aestheticmappingby settingmapping = gg$aes(x = dep_delay, y = arr_delay). Specifically, the variabledep_delaymaps to thexposition aesthetic, while the variablearr_delaymaps to theyposition.
We then add a layer to the ggplot() function call using the + sign.
The added layer in question specifies the third component of the grammar:
the geometric object.
In this case, the geometric object is set to be points
by specifying geom_point().
After running these two lines of code in your console,
you’ll notice two outputs:
a warning message and the graphic shown in Figure 2.2.
Warning: Removed 5 rows containing missing values (geom_point).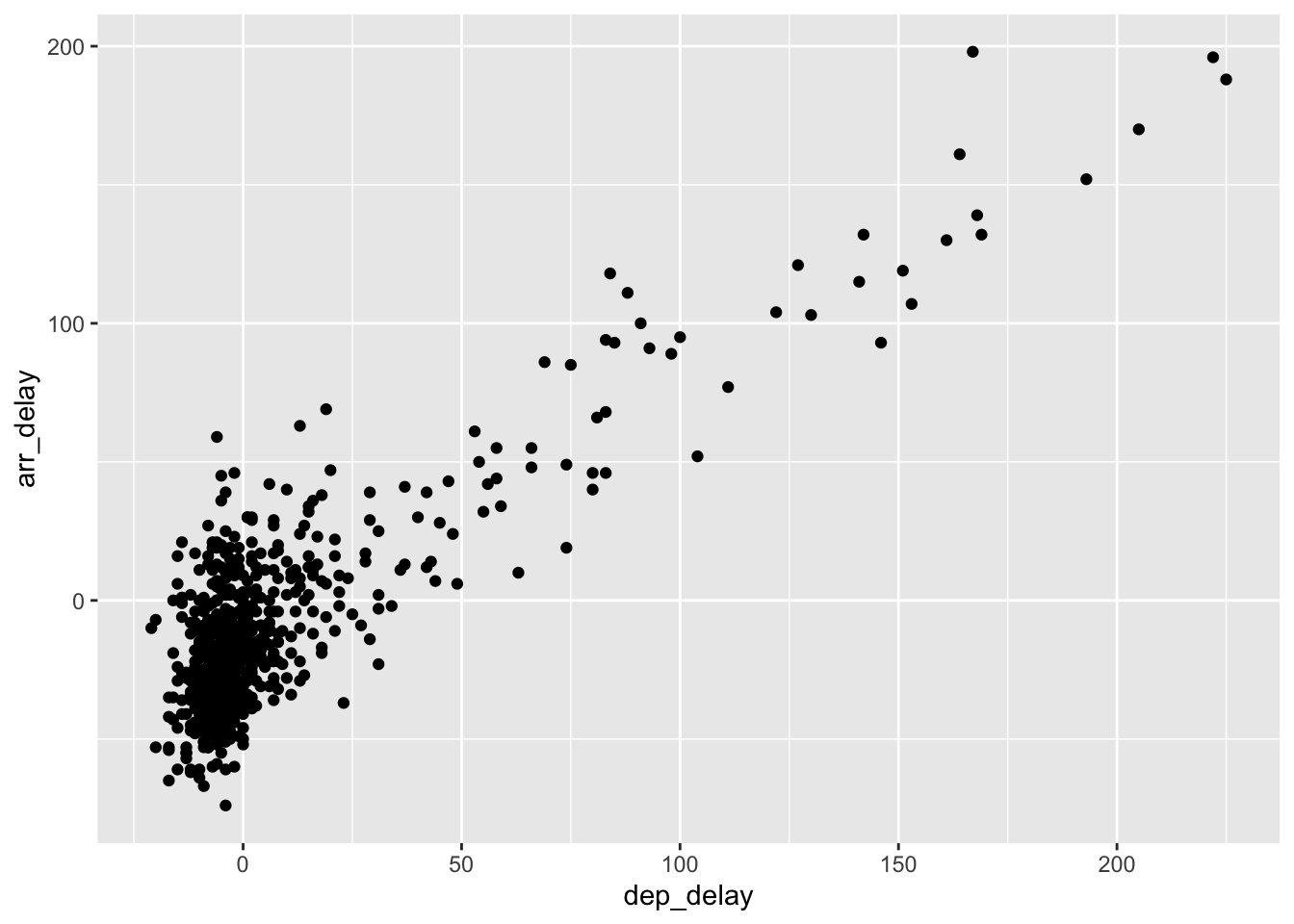
Figure 2.2: Arrival delays versus departure delays for Alaska Airlines flights from NYC in 2013.
Let’s first unpack the graphic in Figure 2.2.
Observe that a positive relationship
exists between dep_delay and arr_delay:
as departure delays increase, arrival delays tend to also increase.
Observe also the large mass of points clustered near (0, 0),
the point indicating flights that neither departed nor arrived late.
Let’s turn our attention to the warning message.
R is alerting us to the fact that five rows were ignored
due to them being missing.
For these 5 rows, either the value for dep_delay or arr_delay
or both were missing (recorded in R as NA),
and thus these rows were ignored in our plot.
Let’s confirm that there are indeed five rows
with missing data by identifying those rows.
| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2013 | 2 | 8 | NA | 1815 | NA | NA | 2130 | NA | AS | 7 | N402AS | EWR | SEA | NA | 2402 | 18 | 15 | 2013-02-08 18:00:00 |
| 2013 | 2 | 9 | NA | 725 | NA | NA | 1035 | NA | AS | 11 | N592AS | EWR | SEA | NA | 2402 | 7 | 25 | 2013-02-09 07:00:00 |
| 2013 | 5 | 6 | 726 | 725 | 1 | 1022 | 1015 | NA | AS | 21 | N407AS | EWR | SEA | NA | 2402 | 7 | 25 | 2013-05-06 07:00:00 |
| 2013 | 5 | 22 | 724 | 725 | -1 | 1135 | 1015 | NA | AS | 21 | N592AS | EWR | SEA | NA | 2402 | 7 | 25 | 2013-05-22 07:00:00 |
| 2013 | 5 | 25 | 724 | 725 | -1 | 1216 | 1015 | NA | AS | 21 | N516AS | EWR | SEA | NA | 2402 | 7 | 25 | 2013-05-25 07:00:00 |
Sure enough, the five row all have missing data in one or the other columns. (Place your mouse cursor inside the table and scroll right to see more columns.)
Before we continue,
let’s make a few more observations about the code
that created the scatterplot.
Note that the + sign comes at the end of lines, and not at the beginning.
You’ll get an error in R if you put it at the beginning of a line.
When adding layers to a plot,
you are encouraged to start a new line after the +
(by pressing the Return/Enter button on your keyboard)
so that the code for each layer is on a new line.
As we add more and more layers to plots,
you’ll see this will greatly improve the legibility of your code.
To stress the importance of adding the layer specifying the geometric object,
consider Figure 2.3 where no layers are added.
Because the geometric object was not specified,
we have a blank plot which is not very useful!
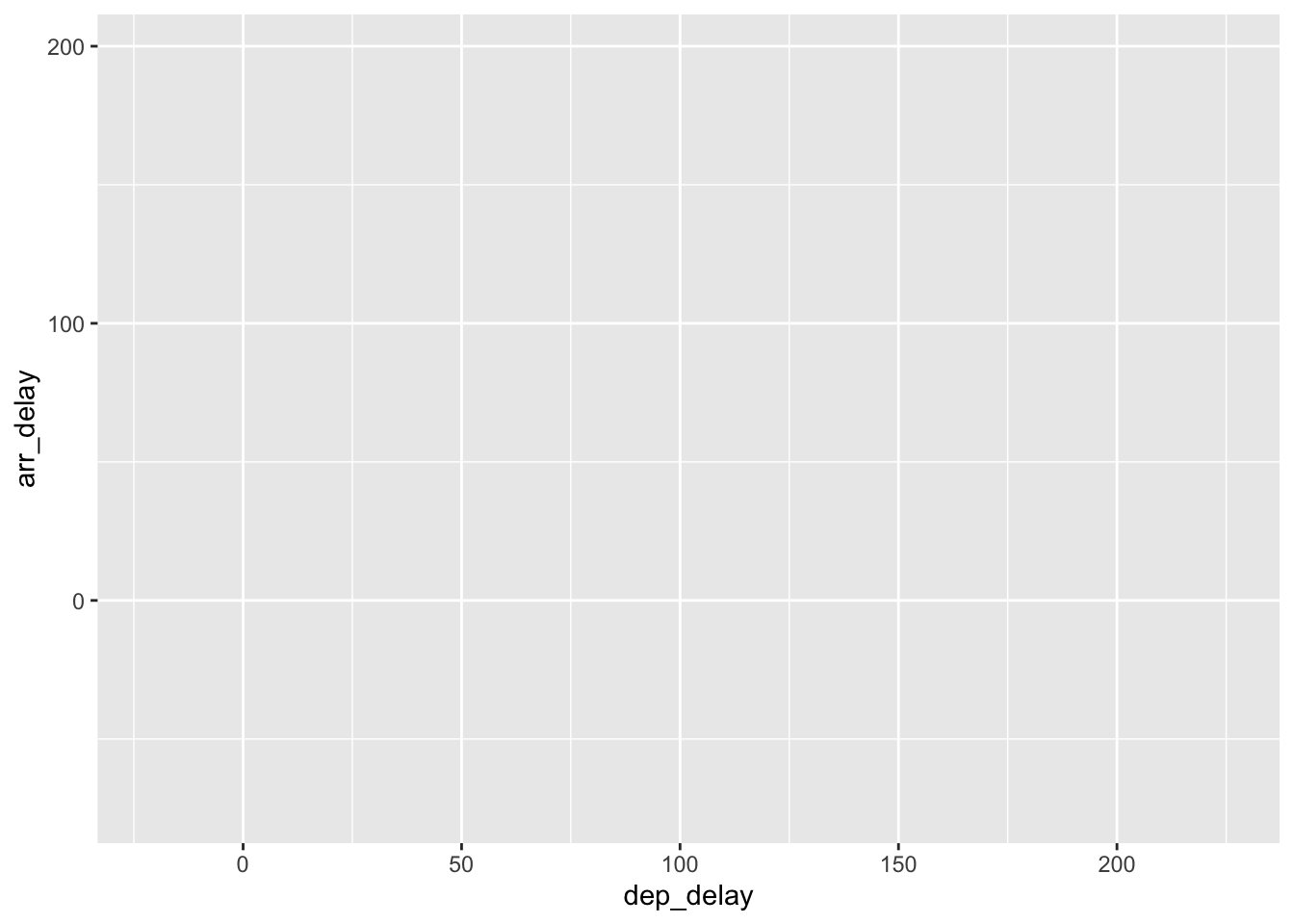
Figure 2.3: A plot with no layers.
Learning check
(LC2.1)
What are some practical reasons why dep_delay and arr_delay
have a positive relationship?
(LC2.2)
What variables in the nycflights13::weather data frame would you expect
to have a negative correlation (i.e., a negative relationship)
with dep_delay? Why?
Remember that we are focusing on continuous variables here.
Hint: Explore the nycflights13::weather dataset by using the View() function.
(LC2.3) Why do you believe there is a cluster of points near (0, 0)? What does (0, 0) correspond to in terms of the Alaska Air flights?
(LC2.4) What are some other features of the plot that stand out to you?
(LC2.5)
Create a new scatterplot using different variables
in the df_alaska_flights data frame by modifying the example given.
2.3.2 Overplotting
The large mass of points near (0, 0) in Figure 2.2 can cause some confusion since it is hard to tell the true number of points that are plotted. This is the result of a phenomenon called overplotting. As one may guess, this corresponds to points being plotted on top of each other over and over again. When overplotting occurs, it is difficult to know the number of points being plotted. There are two methods to address the issue of overplotting. Either by
- Adjusting the transparency of the points or
- Adding a little random “jitter,” or random “nudges,” to each of the points.
Method 1: Changing the transparency
The first way of addressing overplotting is to change the transparency/opacity
of the points by setting the alpha argument in geom_point().
We can change the alpha argument to be any value between 0 and 1,
where 0 sets the points to be 100% transparent
and 1 sets the points to be 100% opaque.
By default, alpha is set to 1.
In other words, if we don’t explicitly set an alpha value,
R will use alpha = 1.
Note how the following code is identical to the code in
Section 2.3 that created the scatterplot with overplotting,
but with alpha = 0.2 added to the geom_point() function:
gg$ggplot(data = df_alaska_flights,
mapping = gg$aes(x = dep_delay, y = arr_delay)) +
gg$geom_point(alpha = 0.2)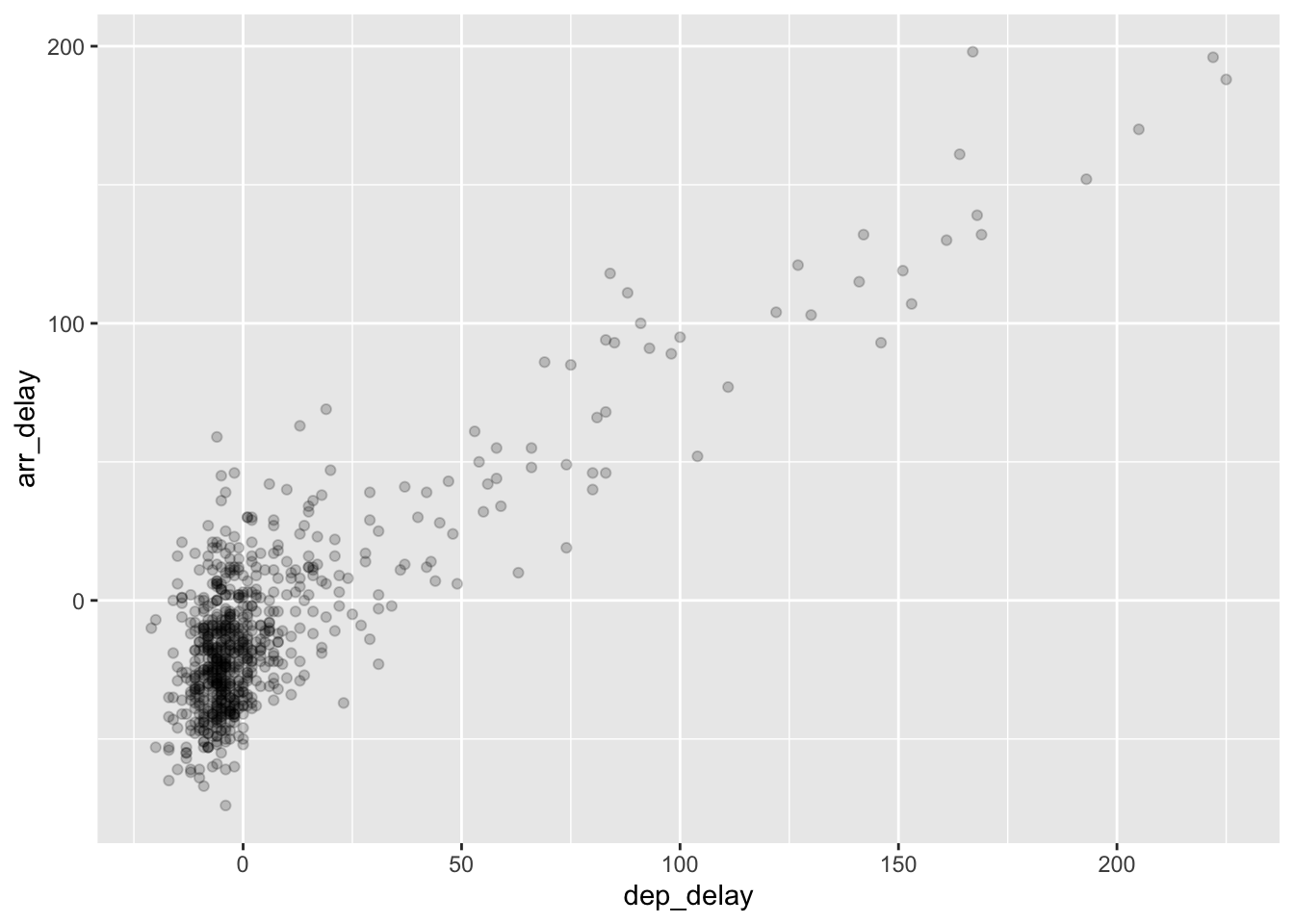
Figure 2.4: Arrival vs. departure delays scatterplot with alpha = 0.2.
The key feature to note in Figure 2.4 is that the transparency of the points is cumulative: areas with a high-degree of overplotting are darker, whereas areas with a lower degree are less dark.
Note furthermore that there is no aes() surrounding alpha = 0.2.
This is because we are not mapping a variable to an aesthetic attribute,
but rather merely changing the default setting of alpha.
In fact, you’ll receive an error if you try to change the second line
to read ggplot2::geom_point(ggplot2::aes(alpha = 0.2)).
Method 2: Jittering the points
The second way of addressing overplotting is by jittering all the points. This means giving each point a small “nudge” in a random direction. You can think of “jittering” as shaking the points around a bit on the plot. Let’s illustrate using a simple example first. Say we have a data frame with 4 identical rows of x and y values: (0,0), (0,0), (0,0), and (0,0). In Figure 2.5, we present both the regular scatterplot of these 4 points (on the left) and its jittered counterpart (on the right).
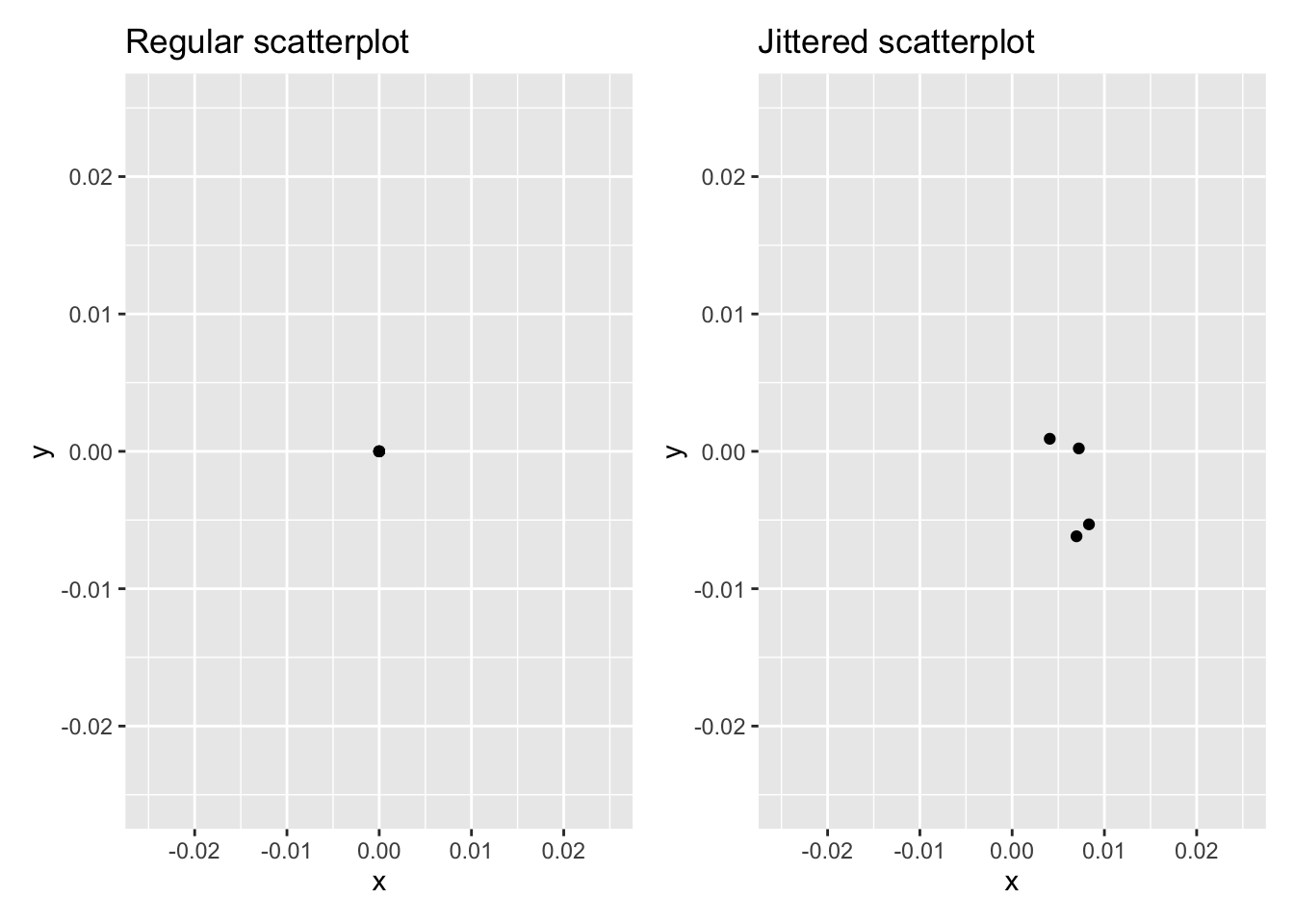
Figure 2.5: Regular and jittered scatterplot.
In the left-hand regular scatterplot, observe that the 4 points are superimposed on top of each other. While we know there are 4 values being plotted, this fact might not be apparent to others. In the right-hand jittered scatterplot, it is now plainly evident that this plot involves four points since each point is given a random “nudge.”
Keep in mind, however, that jittering is strictly a visualization tool; even after creating a jittered scatterplot, the original values saved in the data frame remain unchanged.
To create a jittered scatterplot,
instead of using geom_point(), we use geom_jitter().
Observe how the following code is very similar to the code
that created the scatterplot with overplotting in Subsection 2.3.1,
but with geom_point()
replaced with geom_jitter().
gg$ggplot(data = df_alaska_flights,
mapping = gg$aes(x = dep_delay, y = arr_delay)) +
gg$geom_jitter(width = 30, height = 30)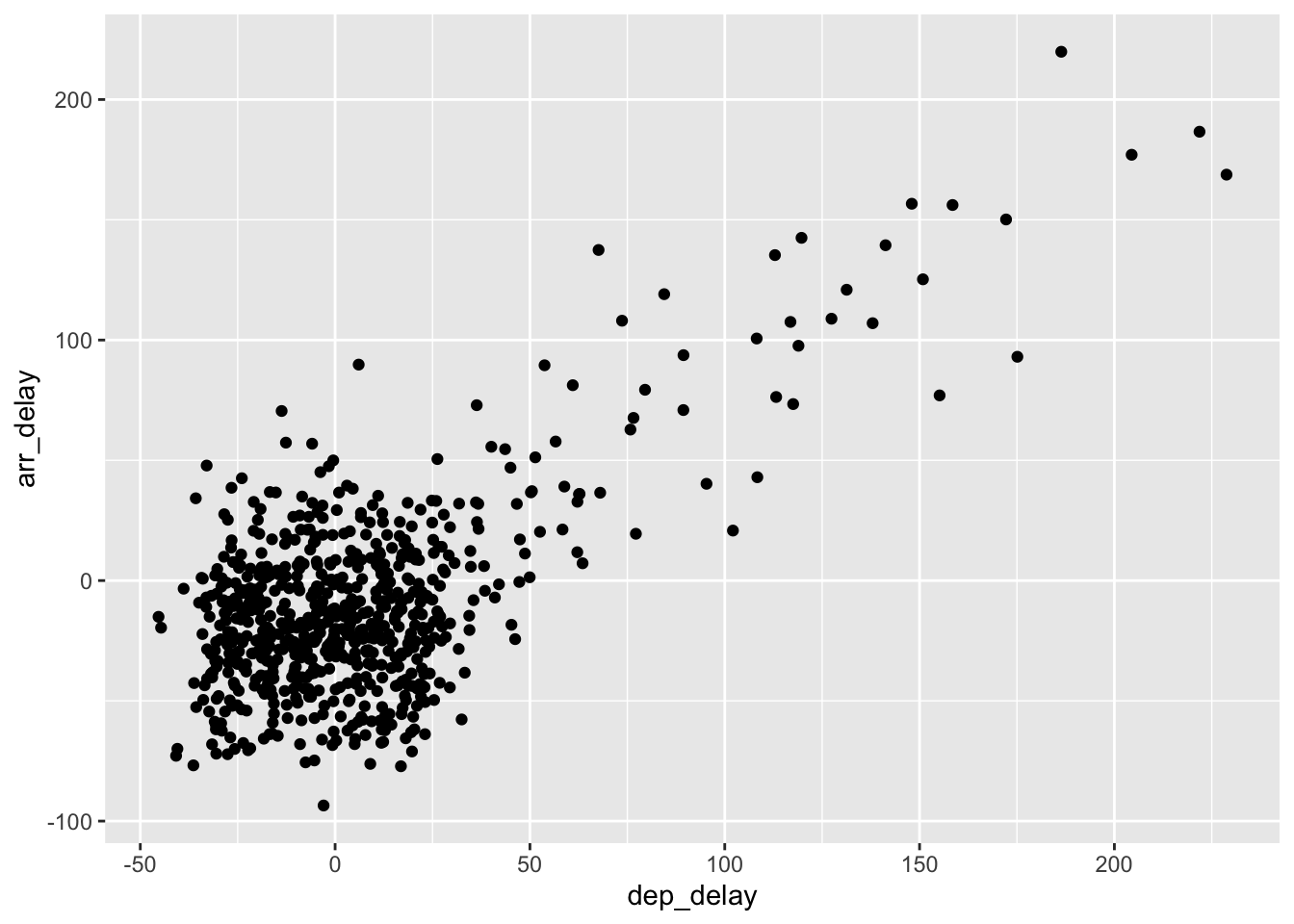
Figure 2.6: Arrival versus departure delays jittered scatterplot.
In order to specify how much jitter to add,
we adjusted the width and height arguments to geom_jitter().
This corresponds to how hard you’d like to shake the plot
in horizontal x-axis units and vertical y-axis units, respectively.
In this case, both axes are in minutes.
How much jitter should we add using the width and height arguments?
On the one hand, it is important to add just enough jitter
to break any overlap in points,
but on the other hand, not so much
that we completely alter the original pattern in points.
As can be seen in the resulting Figure 2.6,
in this case jittering doesn’t really provide much new insight.
In this particular case,
it can be argued that changing the transparency of the points
by setting alpha proved more effective.
When would it be better to use a jittered scatterplot?
When would it be better to alter the points’ transparency?
There is no single right answer that applies to all situations.
You need to make a subjective choice and own that choice.
At the very least when confronted with overplotting, however,
we suggest you make both types of plots
and see which one better emphasizes the point you are trying to make.
Learning check
(LC2.6)
After viewing Figure 2.4,
give an approximate range of arrival delays that occur most frequently.
How about departure delays?
Now compare Figure 2.4 to Figure 2.2.
How has that region changed compared to
when you observed the same plot without alpha = 0.2 set
in Figure 2.2?
(LC2.7)
What additional information does it give you
by setting the alpha argument that a regular scatterplot cannot?
2.3.3 Summary
Scatterplots display the relationship between two continuous variables. They are among the most commonly used plots because they can provide an immediate way to see the trend in one continuous variable versus another. However, if you try to create a scatterplot where either one of the two variables is not continuous, you might get strange results. Be careful!
With medium to large datasets, you may need to play around with the different modifications to scatterplots we saw such as changing the transparency/opacity of the points or by jittering the points. This tweaking is often a fun part of data visualization, since you’ll have the chance to see different relationships emerge as you tinker with your plots.
2.4 5NG#2: Linegraphs
The next of the five named graphs are linegraphs. Linegraphs show the relationship between two continuous variables when the variable on the x-axis, also called the explanatory variable, is of a sequential nature. In other words, there is an inherent ordering to the variable.
The most common examples of linegraphs have some notion of time
on the x-axis: hours, days, weeks, years, etc.
Since time is sequential,
we connect consecutive observations of the variable on the y-axis with a line.
Linegraphs that have some notion of time on the x-axis are also called
time series plots.
Let’s illustrate linegraphs using another dataset in the
nycflights13 package:
the weather data frame.
Let’s explore the weather data frame
by running View(nycflights13::weather) or glimpse(nycflights13::weather).
Furthermore let’s read the associated help file
by running ?nycflights13::weather to bring up the help file.
Observe that there is a variable called temp
of hourly temperature recordings in Fahrenheit at weather stations
near all three major airports in New York City:
Newark (origin code EWR),
John F. Kennedy International (JFK),
and LaGuardia (LGA).
However, instead of considering hourly temperatures for all days in 2013
for all three airports,
for simplicity let’s only consider hourly temperatures
at Newark airport for the first 15 days in January.
Recall in Section 2.3,
we used the filter() function to only choose the subset of rows of flights
corresponding to Alaska Airlines flights.
We similarly use filter() here,
but by using the & operator
we only choose the subset of rows of weather where the origin is "EWR",
the month is January, and the day is between 1 and 15.
Recall we performed a similar task in Section 2.3
when creating the df_alaska_flights data frame of only Alaska Airlines flights,
a topic we’ll explore more in Chapter 3 on data wrangling.
Before applying filter,
let’s import the weather dataset from package nycflights13,
so it can be referred to in the short form df_weather.
Furthermore, let’s convert Fahrenheit to Celsius.
# import the data "weather" and save it as "df_weather"
import::from(nycflights13, df_weather = weather)
# convert Fahrenheit to Celsius
df_weather$temp_c = (df_weather$temp - 32) * (5 / 9)# Reduce the data to the intended subset
early_january_weather <- df_weather %>%
dp$filter(origin == "EWR" & month == 1 & day <= 15)Learning check
(LC2.8)
Take a look at both the df_weather and
early_january_weather data frames
by running View(df_weather) and View(early_january_weather).
In what respect do these data frames differ?
(LC2.9)
View() the df_flights data frame again.
Why does the time_hour variable uniquely identify the hour of the measurement,
whereas the hour variable does not?
2.4.1 Linegraphs via geom_line
Let’s create a time series plot of the hourly temperatures
saved in the early_january_weather data frame
by using geom_line() to create a linegraph,
instead of geom_point() which we used previously to create scatterplots:
gg$ggplot(data = early_january_weather,
mapping = gg$aes(x = time_hour, y = temp_c)) +
gg$geom_line() +
gg$labs(y = "Temperature (Celsius)")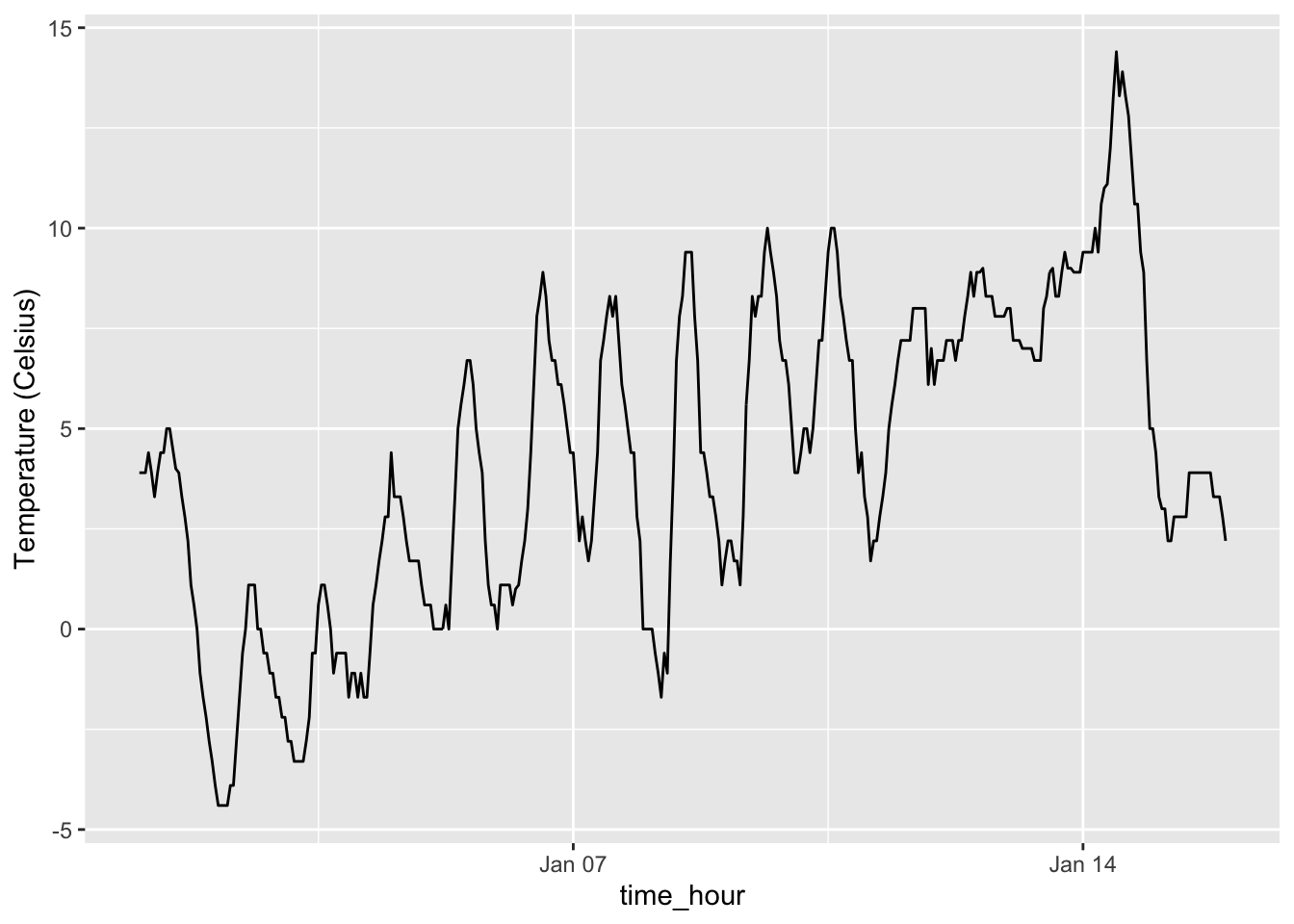
Figure 2.7: Hourly temperature in Newark for January 1-15, 2013.
Similar to the ggplot() code that created the scatterplot of departure
and arrival delays for Alaska Airlines flights in Figure 2.2,
let’s break down this code piece-by-piece in terms of the grammar of graphics:
Within the ggplot() function call,
we specify two of the components of the grammar of graphics as arguments:
- The
datato be theearly_january_weatherdata frame by settingdata = early_january_weather. - The
aestheticmappingby settingmapping = gg$aes(x = time_hour, y = temp_c). Specifically, the variabletime_hourmaps to thexposition aesthetic, while the variabletemp_cmaps to theyposition aesthetic.
We add a layer to the ggplot() function call using the + sign.
The layer in question specifies the third component of the grammar:
the geometric object in question.
In this case, the geometric object is a line set by specifying geom_line().
Learning check
(LC2.10)
Plot a time series of temp_c for Newark Airport
in the first 15 days of June 2013.
(LC2.11) Why should linegraphs be avoided when there is not a clear ordering of the horizontal axis?
2.5 5NG#3: Histograms
Let’s consider the temp_c variable in the df_weather data frame once again,
but unlike with the linegraphs in Section 2.4,
let’s say we don’t care about its relationship with time,
but rather we only care about how the values of temp_c distribute.
In other words:
- What are the smallest and largest values?
- What is the “center” or “most typical” value?
- How do the values spread out?
- What are frequent and infrequent values?
One way to visualize this distribution
of this single variable temp_c is to plot them on a horizontal line:

Figure 2.8: Plot of hourly temperature recordings from NYC in 2013.
This gives us a general idea of how the values of temp_c distribute:
observe that temperatures vary from around
-12°C
up to 38°C.
Furthermore, there appear to be more recorded temperatures
between 5°C and 15°C than outside this range.
However, because of the high degree of overplotting in the points,
it’s hard to get a sense of exactly how many values
are between say 5°C and 10°C.
What is commonly produced instead of Figure 2.8
is known as a histogram.
A histogram is a plot that visualizes the distribution
of a continuous value as follows:
- We first cut up the x-axis into a series of bins, where each bin represents a range of values.
- For each bin, we count the number of observations that fall in the range corresponding to that bin.
- Then for each bin, we draw a bar whose height marks the corresponding count.
Let’s drill-down on an example of a histogram, shown in Figure 2.9.
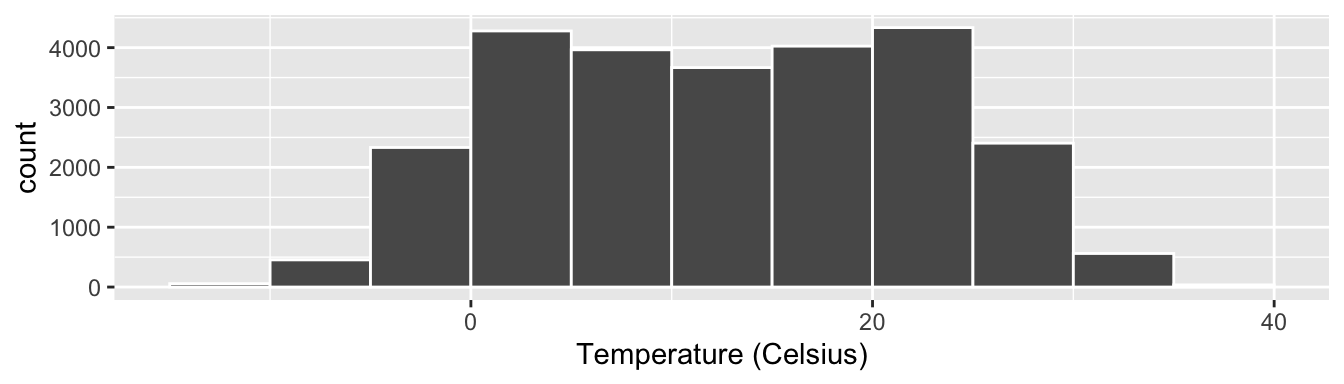
Figure 2.9: Example histogram.
Let’s focus only on temperatures between 0°C and 20°C for now. Observe that there are four bins of equal width between 0°C and 20°C. Thus we have four bins of width 5°C each: one bin for the 0-5°C range, another bin for the 5-10°C range, and another bin for the 10-15°C range. Since:
- The bin for the 5-10°C range has a height of around 4000. In other words, around 4000 of the hourly temperature recordings are between 5°C and 10°C.
- The bin for the 10-15°C range has a height of around 3500. In other words, around 3500 of the hourly temperature recordings are between 10°C and 15°C.
All nine bins spanning -10°C to 35°C on the x-axis have this interpretation.
2.5.1 Histograms via geom_histogram
Let’s now present the ggplot() code to plot your first histogram!
Unlike with scatterplots and linegraphs,
there is now only one variable being mapped in aes():
the single continuous variable temp_c.
The y-aesthetic of a histogram, the count of the observations in each bin,
gets computed for you automatically.
Furthermore, the geometric object layer is now a geom_histogram().
After running the following code,
you’ll see the histogram in Figure 2.10
as well as warning messages. We’ll discuss the warning messages first.
gg$ggplot(data = df_weather,
mapping = gg$aes(x = temp_c)) +
gg$geom_histogram() +
gg$labs(x = "Temperature (Celsius)")`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 1 rows containing non-finite values (stat_bin).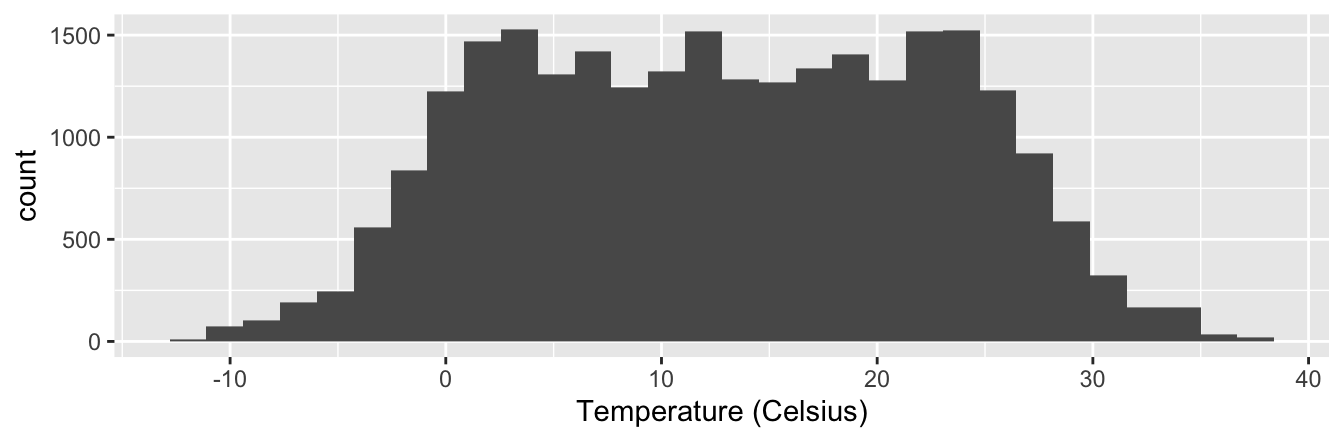
Figure 2.10: Histogram of hourly temperatures at three NYC airports.
The first warning message is telling us that the histogram was constructed using bins = 30 for 30 equally spaced bins. This is known in computer programming as a default value; unless you override this default number of bins with a number you specify, R will choose 30 by default. We’ll see in the next section how to change the number of bins to another value than the default.
The second message is telling us something similar to the warning message we received when we ran the code to create a scatterplot of departure and arrival delays for Alaska Airlines flights in Figure 2.2: that because one row has a missing NA value for temp_c, it was omitted from the histogram. R is just giving us a friendly heads up that this was the case.
Now let’s unpack the resulting histogram in Figure 2.10.
Observe that values less than -5°C as well as values above 35°C
are rather rare.
However, because of the large number of bins,
it’s hard to get a sense for which range of temperatures is spanned by each bin;
everything is one giant amorphous blob.
So let’s add white vertical borders demarcating the bins
by adding a color = "white" argument to geom_histogram()
and ignore the warning about setting the number of bins to a better value:
gg$ggplot(data = df_weather,
mapping = gg$aes(x = temp_c)) +
gg$geom_histogram(color = "white") +
gg$labs(x = "Temperature (Celsius)")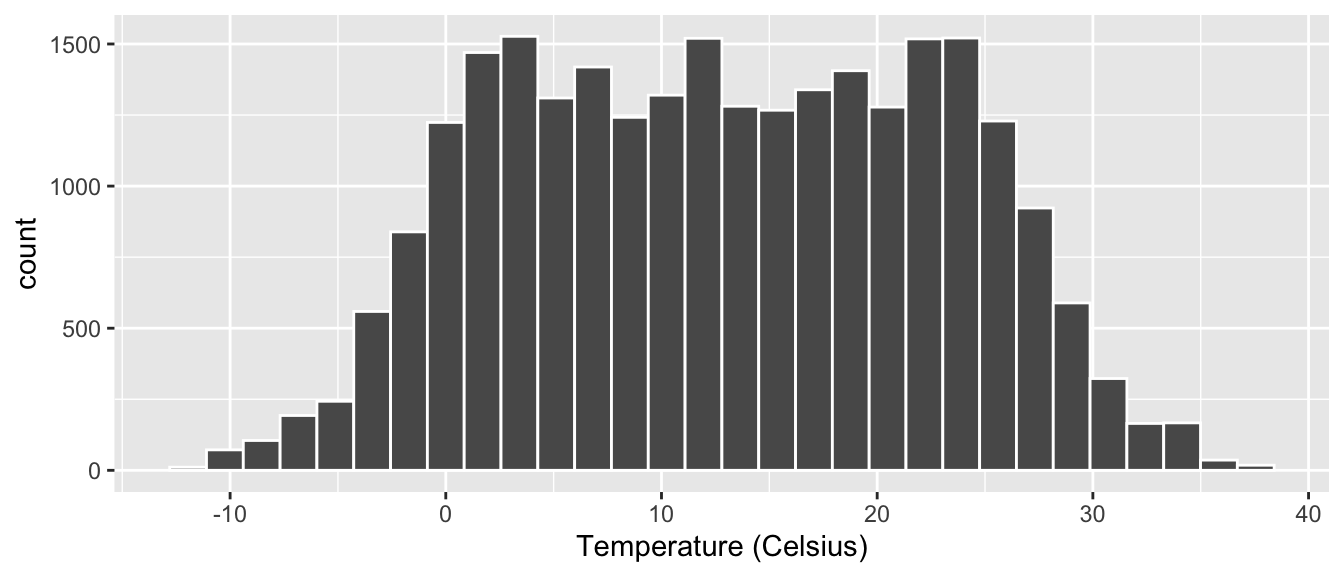
Figure 2.11: Histogram of hourly temperatures at three NYC airports with white borders.
We now have an easier time associating ranges of temperatures
to each of the bins in Figure 2.11.
We can also vary the color of the bars
by setting the fill argument.
For example, you can set the bin colors to be “blue steel”
by setting fill = "steelblue":
# Try on your own
gg$ggplot(data = df_weather,
mapping = gg$aes(x = temp_c)) +
gg$geom_histogram(color = "white", fill = "steelblue") +
gg$labs(x = "Temperature (Celsius)")If you’re curious, run colors() to see
all 657 possible choice of colors in R!
2.5.2 Adjusting the bins
Observe in Figure 2.11 that in the 0-10°C range there appear to be 6 bins. Thus each bin has a width of 10 divided by 6, or 1.667°C, which is not a very easily interpretable range to work with. Let’s improve this by adjusting the number of bins in our histogram in one of two ways:
- By adjusting the number of bins via the
binsargument ingeom_histogram(). - By adjusting the width of the bins via the
binwidthargument ingeom_histogram().
Using the first method, we have the power to specify how many bins we would like to cut the x-axis up in. As mentioned in the previous section, the default number of bins is 30. We can override this default, to say 40 bins, as follows:
gg$ggplot(data = df_weather,
mapping = gg$aes(x = temp_c)) +
gg$geom_histogram(bins = 40, color = "white") +
gg$labs(x = "Temperature (Celsius)")Using the second method,
instead of specifying the number of bins,
we specify the width of the bins by using the binwidth argument
in the geom_histogram() layer.
For example, let’s set the width of each bin to be 5°C.
gg$ggplot(data = df_weather,
mapping = gg$aes(x = temp_c)) +
gg$geom_histogram(binwidth = 5, color = "white") +
gg$labs(x = "Temperature (Celsius)")We compare both resulting histograms side-by-side in Figure 2.12.
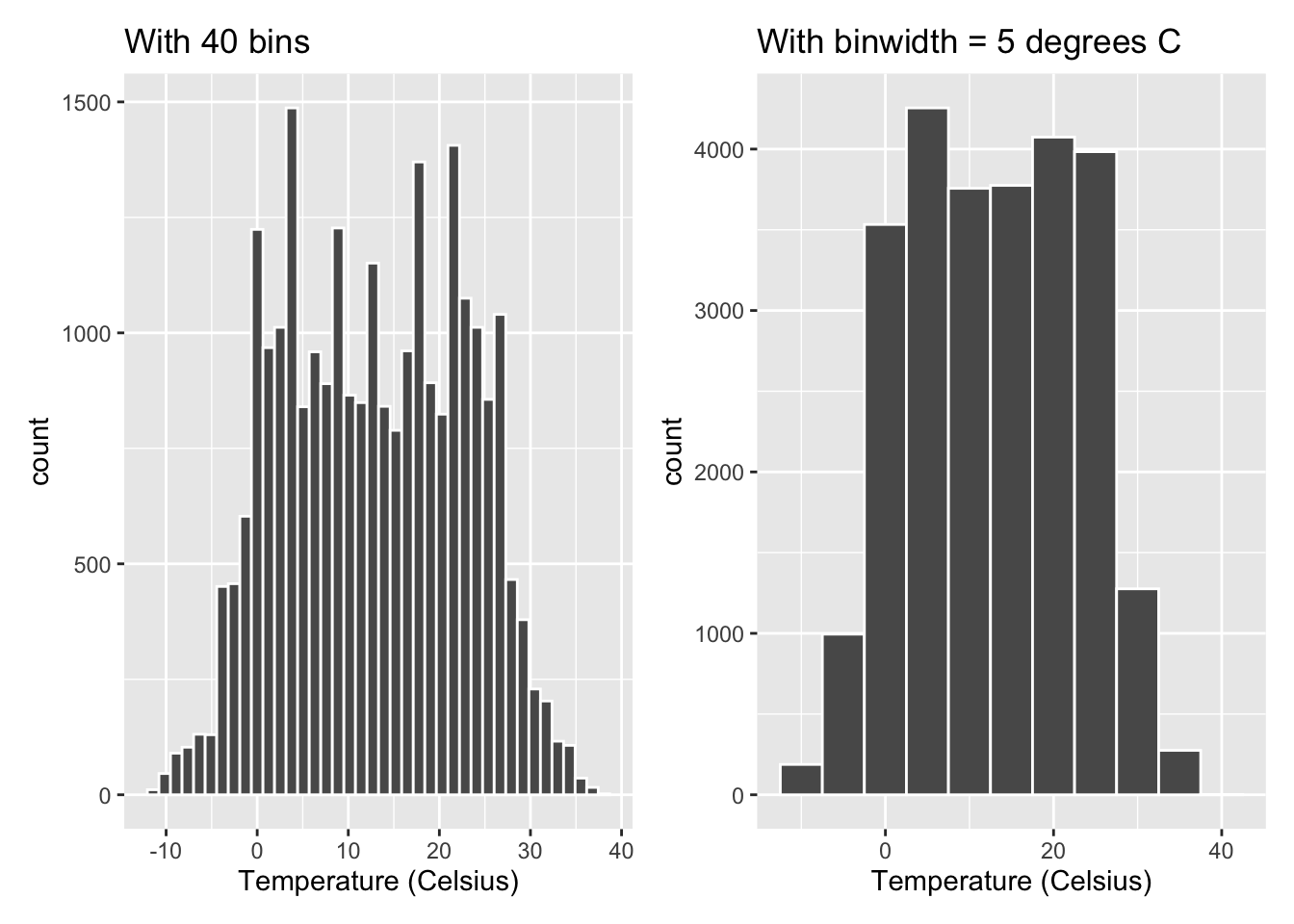
Figure 2.12: Setting histogram bins in two ways.
Learning check
(LC2.12) Change the number of bins to 20 and redraw the plot. How is the plot different from when the number of bins is 30? 40? What can you conclude about how to choose the number of bins?
(LC2.13) Would you classify the distribution of temperatures as symmetric or skewed in one direction or another?
(LC2.14)
What would you guess is the “center” value in this distribution?
Why did you make that choice?
(LC2.15) Is this data spread out greatly from the center or is it close? Why?
2.6 Facets
Before continuing with the next of the 5NG,
let’s briefly introduce a new concept called faceting.
Faceting is used when we’d like to split a particular visualization
by the values of another variable.
This will create multiple copies of the same type of plot
with matching x and y axes, but whose content will differ.
For example, suppose we were interested in looking at
how the histogram of hourly temperature recordings at the three NYC airports
we saw in Figure 2.9 differed in each month.
We could “split” this histogram by the 12 possible months in a given year.
In other words, we would plot histograms of temp_c for each month separately.
We do this by adding facet_wrap(~ month) layer.
Note the ~ is a “tilde” and you can type it
by pressing shift and the key next to the “1” key on most US keyboards.
The tilde is required and you’ll receive the error
Error in as.quoted(facets) : object 'month' not found
if you don’t include it here.
gg$ggplot(data = df_weather,
mapping = gg$aes(x = temp_c)) +
gg$geom_histogram(binwidth = 5, color = "white") +
gg$facet_wrap(~ month) +
gg$labs(x = "Temperature (Celsius)")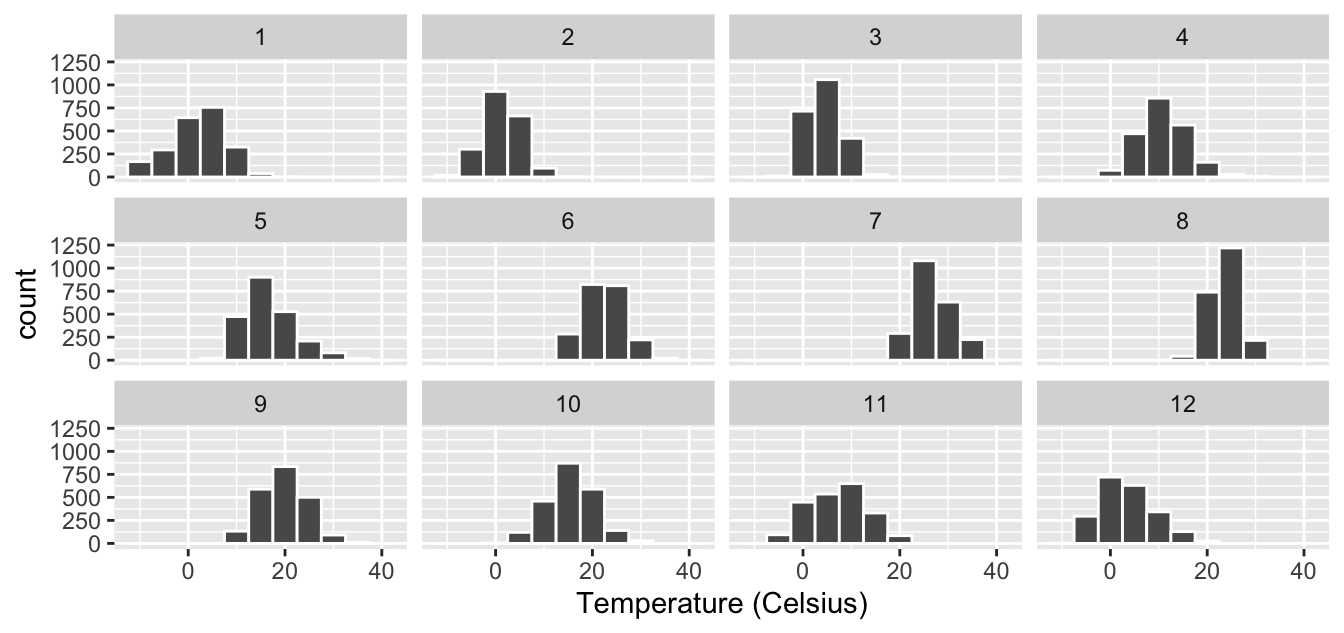
Figure 2.13: Faceted histogram of hourly temperatures by month.
We can also specify the number of rows and columns in the grid
by using the nrow and ncol arguments
inside of facet_wrap().
For example, say we would like our faceted histogram
to have 4 rows instead of 3.
We simply add an nrow = 4 argument to facet_wrap(~ month).
gg$ggplot(data = df_weather,
mapping = gg$aes(x = temp_c)) +
gg$geom_histogram(binwidth = 5, color = "white") +
gg$facet_wrap(~ month, nrow = 4) +
gg$labs(x = "Temperature (Celsius)")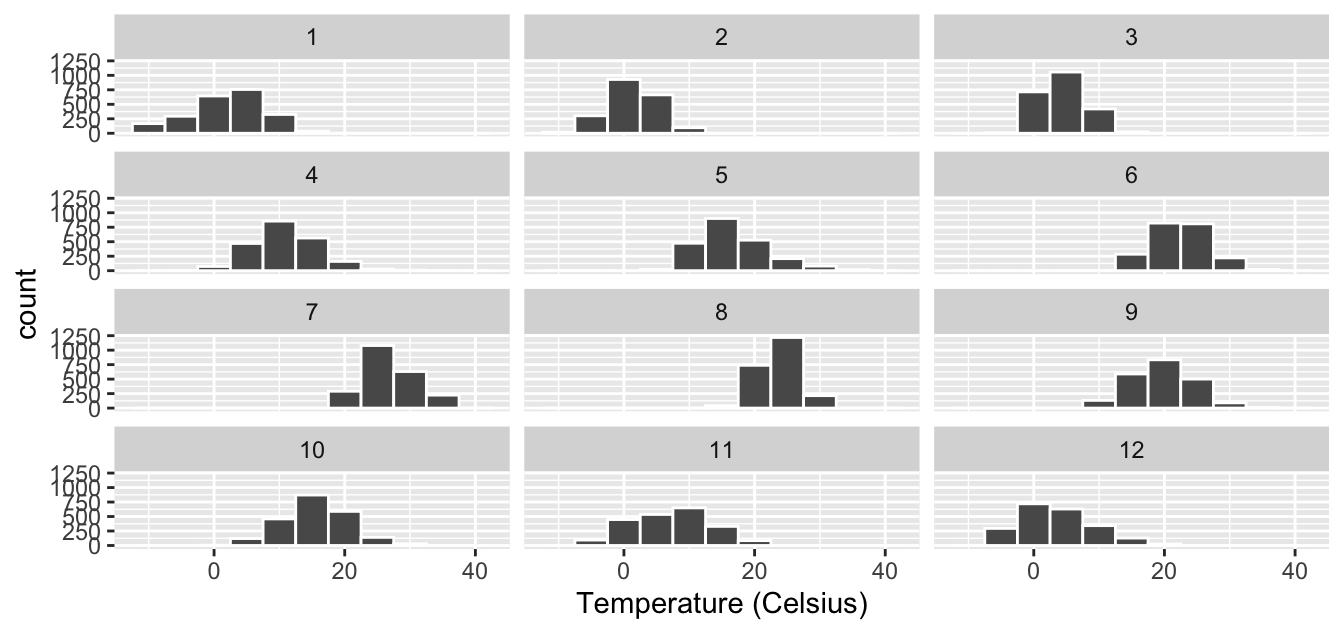
Figure 2.14: Faceted histogram with 4 instead of 3 rows.
Observe in both Figures 2.13 and 2.14 that as we might expect in the Northern Hemisphere, temperatures tend to be higher in the summer months, while they tend to be lower in the winter.
Learning check
(LC2.16) What other things do you notice about this faceted plot? How does a faceted plot help us see relationships between two variables?
(LC2.17) What do the numbers 1-12 correspond to in the plot? What about 0, 20, 40?
(LC2.18) For which types of datasets would faceted plots not work well? Give an example of such a dataset and describe some of its important characteristics.
2.7 5NG#4: Boxplots
Faceted histograms are one type of visualization used to compare the distribution of a continuous variable split by the values of another variable. Another type of visualization that achieves this same goal is a side-by-side boxplot. A boxplot is constructed from the information provided in the five-number summary of a continuous variable (see Appendix A.1).
To keep things simple for now, let’s only consider the 2141 hourly temperature recordings for the month of November, each represented as a jittered point in Figure 2.15.
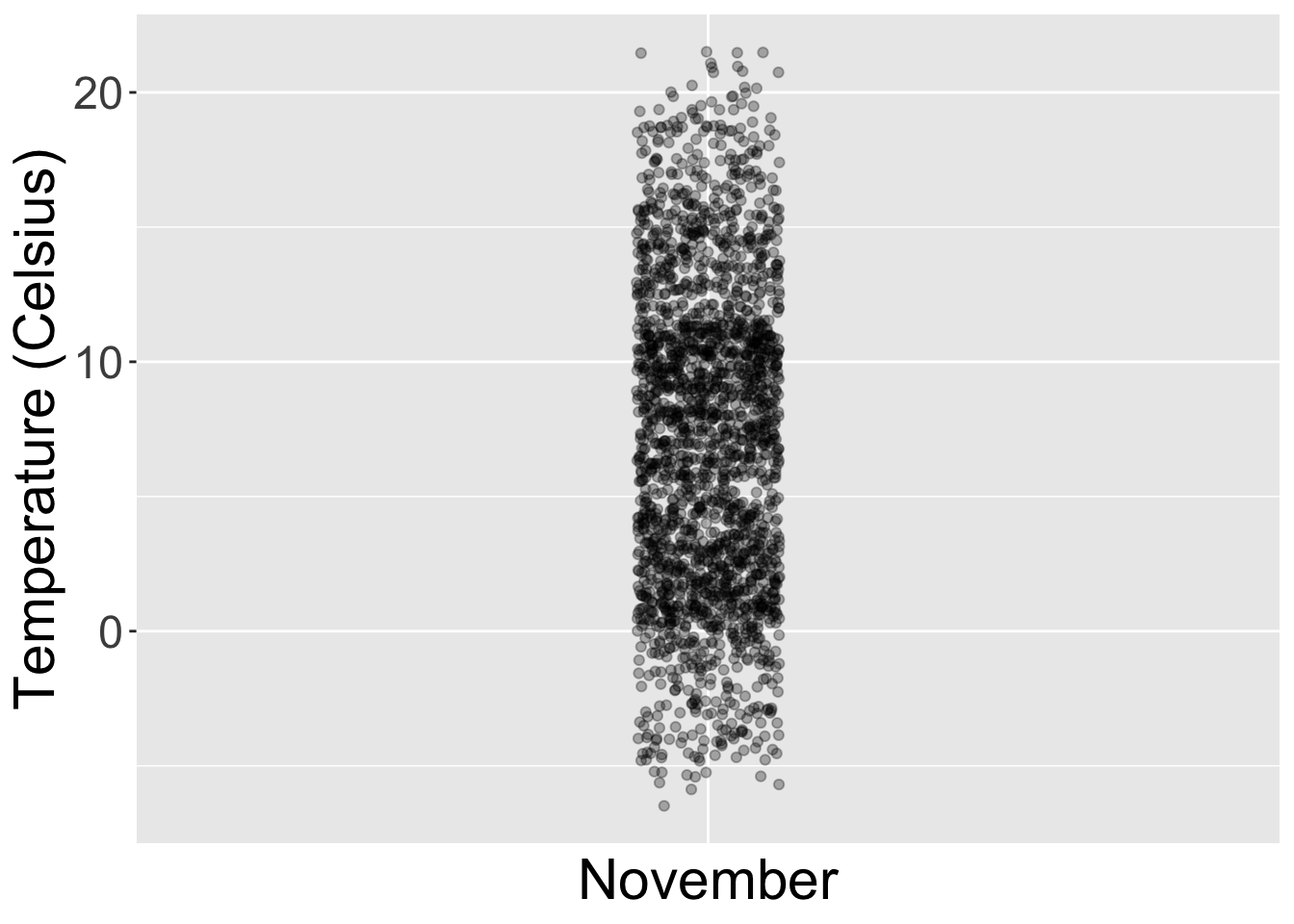
Figure 2.15: November temperatures represented as jittered points.
These 2141 observations have the following five-number summary:
- Minimum: -6°C
- First quartile (25th percentile): 2°C
- Median (second quartile, 50th percentile): 7°C
- Third quartile (75th percentile): 11°C
- Maximum: 22°C
In the leftmost plot of Figure 2.16, let’s mark these 5 values with dashed horizontal lines on top of the 2141 points. In the middle plot of Figure 2.16 let’s add the boxplot. In the rightmost plot of Figure 2.16, let’s remove the points and the dashed horizontal lines for clarity’s sake.
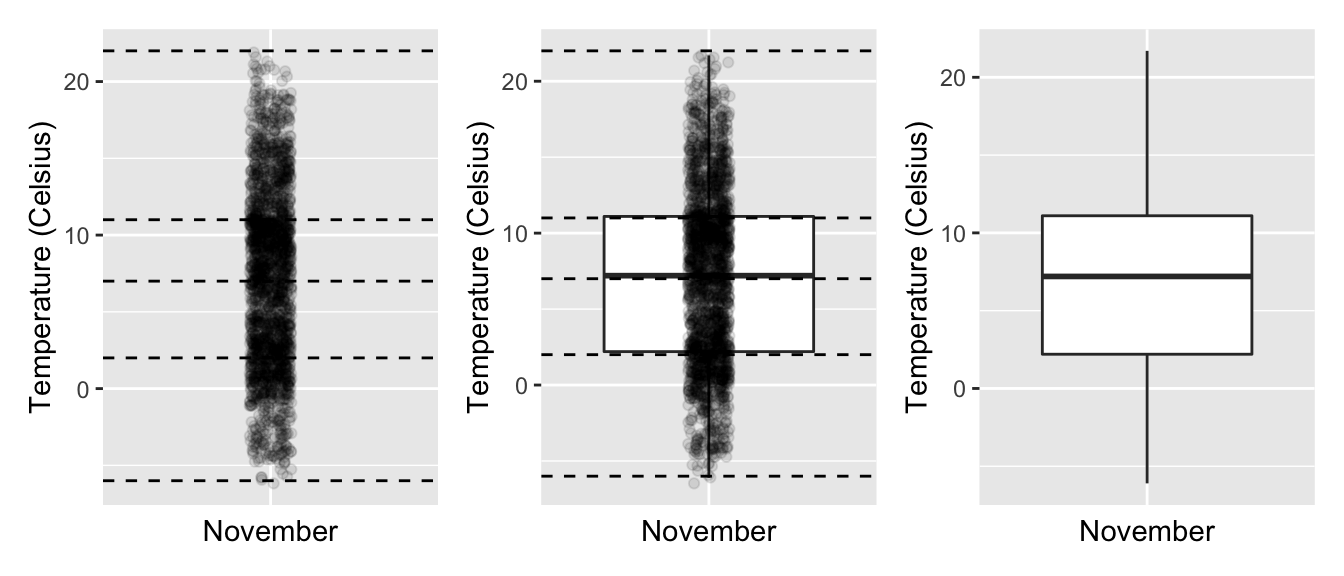
Figure 2.16: Building up a boxplot of November temperatures.
What the boxplot does is visually summarize the 2141 points by cutting the 2141 temperature recordings into quartiles at the dashed lines, where each quartile contains roughly 2141 4 535 observations. Thus
- 25% of points fall below the bottom edge of the box, which is the first quartile of 2°C. In other words, 25% of observations were below 2°C.
- 25% of points fall between the bottom edge of the box and the solid middle line, which is the median of 7°C. Thus, 25% of observations were between 2°C and 7°C and 50% of observations were below 7°C.
- 25% of points fall between the solid middle line and the top edge of the box, which is the third quartile of 11°C. It follows that 25% of observations were between 7°C and 11°C and 75% of observations were below 11°C.
- 25% of points fall above the top edge of the box. In other words, 25% of observations were above 11°C.
- The middle 50% of points lie within the interquartile range (IQR) between the first and third quartile. Thus, the IQR for this example is 11 - 2 = 9°C. The interquartile range is a measure of a continuous variable’s spread.
Furthermore, in the rightmost plot of Figure 2.16, we see the whiskers of the boxplot. The whiskers stick out from either end of the box all the way to the minimum and maximum observed temperatures of -6°C and 22°C, respectively. However, the whiskers don’t always extend to the smallest and largest observed values as they do here. They in fact extend no more than 1.5 the interquartile range from either end of the box. In this case of the November temperatures, no more than 1.5 9°C = 13.5°C from either end of the box. Any observed values outside this range get marked with points called outliers, which we’ll see in the next section.
2.7.1 Boxplots via geom_boxplot
Let’s now create a side-by-side boxplot
of hourly temperatures split by the 12 months
as we did previously with the faceted histograms.
We do this by mapping the month variable to the x-position aesthetic,
the temp_c variable to the y-position aesthetic,
and by adding a geom_boxplot() layer:
gg$ggplot(data = df_weather,
mapping = gg$aes(x = factor(month, ordered=T), y = temp_c)) +
gg$geom_boxplot() +
gg$labs(x = "Month", y = "Temperature (Celsius)")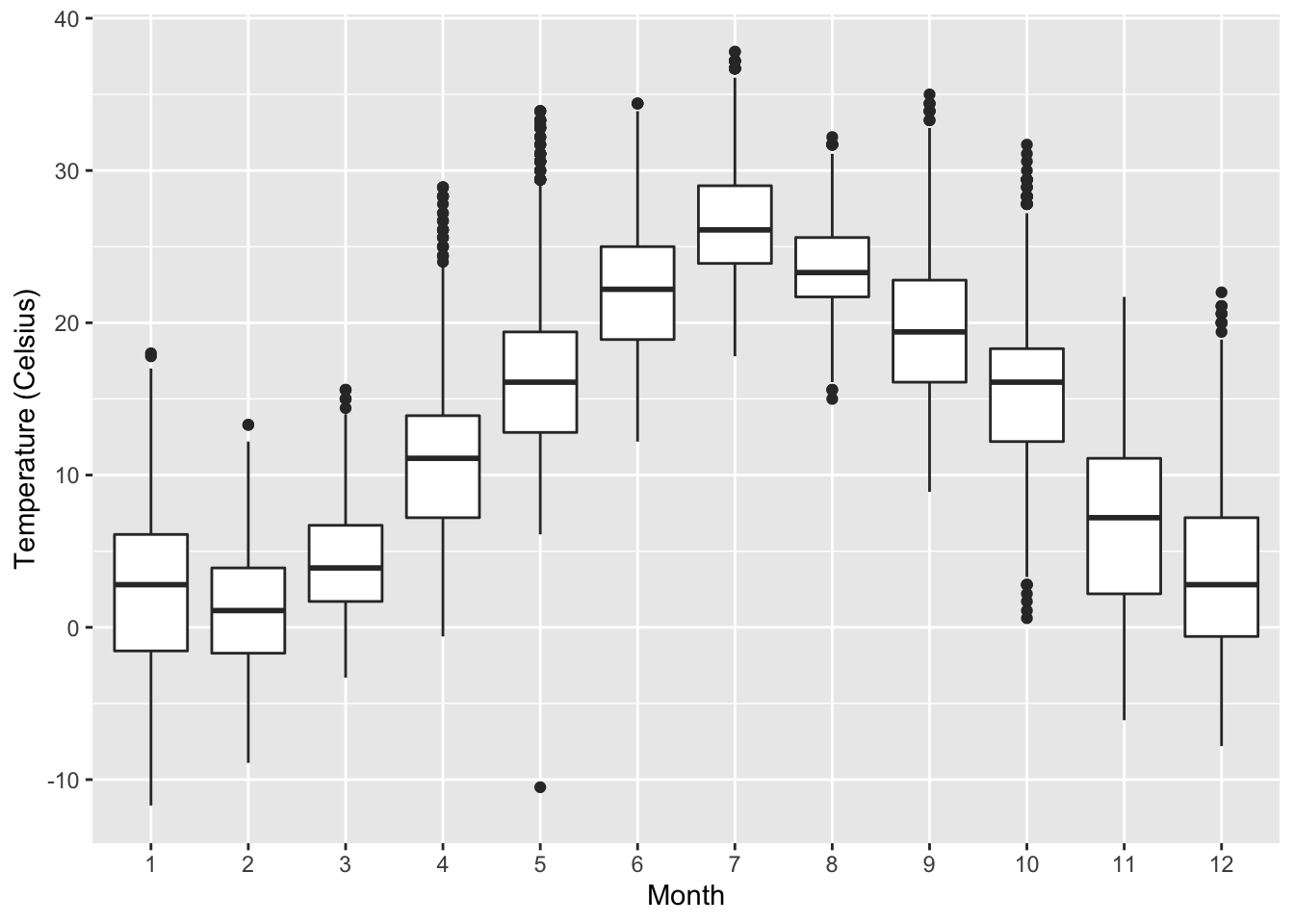
Figure 2.17: Side-by-side boxplot of temperature split by month.
The code above shares the same structure as what we have seen
with scatterplots and linegraphs,
with one distinguished feature: x = factor(...).
We will discuss this distinction in section 2.10.1.
For now, let’s focus on the main theme: boxplot.
The resulting Figure 2.17 shows 12 separate “box and whiskers” plots similar to the rightmost plot of Figure 2.16 of only November temperatures. Thus the different boxplots are shown “side-by-side.”
- The “box” portions of the visualization represent the 1st quartile, the median (the 2nd quartile), and the 3rd quartile.
- The height of each box (the value of the 3rd quartile minus the value of the 1st quartile) is the interquartile range (IQR). It is a measure of the spread of the middle 50% of values, with longer boxes indicating more variability.
- The “whisker” portions of these plots extend out from the bottoms
and tops of the boxes and represent points less than the 25th percentile
and greater than the 75th percentiles, respectively.
They’re set to extend out no more than units away
from either end of the boxes.
We say “no more than” because the ends of the whiskers
have to correspond to observed temperatures.
The length of these whiskers show how the data outside the middle 50% of values vary, with longer whiskers indicating more variability. - The dots representing values falling outside the whiskers are called outliers. These can be thought of as anomalous (“out-of-the-ordinary”) values.
It is important to keep in mind that the definition of an outlier is somewhat arbitrary and not absolute. In this case, they are defined by the length of the whiskers, which are no more than units long for each boxplot. Looking at this side-by-side plot we can see, as expected, that summer months (6 through 8) have higher median temperatures as evidenced by the higher solid lines in the middle of the boxes. We can easily compare temperatures across months by drawing imaginary horizontal lines across the plot. Furthermore, the heights of the 12 boxes as quantified by the interquartile ranges are informative too; they tell us about variability, or spread, of temperatures recorded in a given month.
Learning check
(LC2.19)
What does the dot at the bottom of the plot for May correspond to?
Explain what might have occurred in May to produce this point.
(LC2.20)
Which months have the highest variability in temperature?
What reasons can you give for this?
(LC2.21)
Boxplots provide a simple way to identify outliers.
Why may outliers be easier to identify
when looking at a boxplot instead of a faceted histogram?
2.7.2 Summary
Side-by-side boxplots provide us with a way to compare the distribution of a continuous variable, such as temperature, across multiple values of another categorical variable. One can see where the median falls across the different groups by comparing the solid lines in the center of the boxes.
To study the spread of a continuous variable within one of the boxes, look at both the length of the box and also how far the whiskers extend from either end of the box. Outliers are even more easily identified when looking at a boxplot than when looking at a histogram as they are marked with distinct points.
2.8 5NG#5: Barplots
A boxplot is often used to visualize the distribution of continuous variables. whereas a barplot is used to visualize the distribution of a categorical variable. Take sex as a categorical variable for example. We can visualize the number of male vs. female studuents in this class. To do so, we begin by counting heads in each category of sex, namely male and female. These categories are also known as the levels of the categorical variable. And the counts of each level are known as frequencies. A barplot is sometimes referred to as a barchart.
Barplots are easy to construct. One complication, however, is how your data is represented. Is the categorical variable of interest “pre-counted” or not? For example, run the following code that manually creates two data frames representing a collection of fruit: 3 apples and 2 oranges.
fruits <- tibble::tibble(
fruit = c("apple", "apple", "orange", "apple", "orange")
)
fruits_counted <- tibble::tibble(
fruit = c("apple", "orange"),
number = c(3, 2)
)We see both the fruits and fruits_counted data frames
represent the same collection of fruit.
Whereas fruits just lists the fruit individually…
# A tibble: 5 x 1
fruit
<chr>
1 apple
2 apple
3 orange
4 apple
5 orange… fruits_counted has a variable count which represent the “pre-counted” values of each fruit.
# A tibble: 2 x 2
fruit number
<chr> <dbl>
1 apple 3
2 orange 2Depending on how your categorical data is represented,
you’ll need to add a different geometric layer type to your ggplot()
to create a barplot, as we now explore.
2.8.1 Barplots via geom_bar or geom_col
Let’s generate barplots using these two different representations
of the same basket of fruit: 3 apples and 2 oranges.
Using the fruits data frame
where all 5 fruits are listed individually in 5 rows,
we map the fruit variable to the x-position aesthetic
and add a geom_bar() layer:
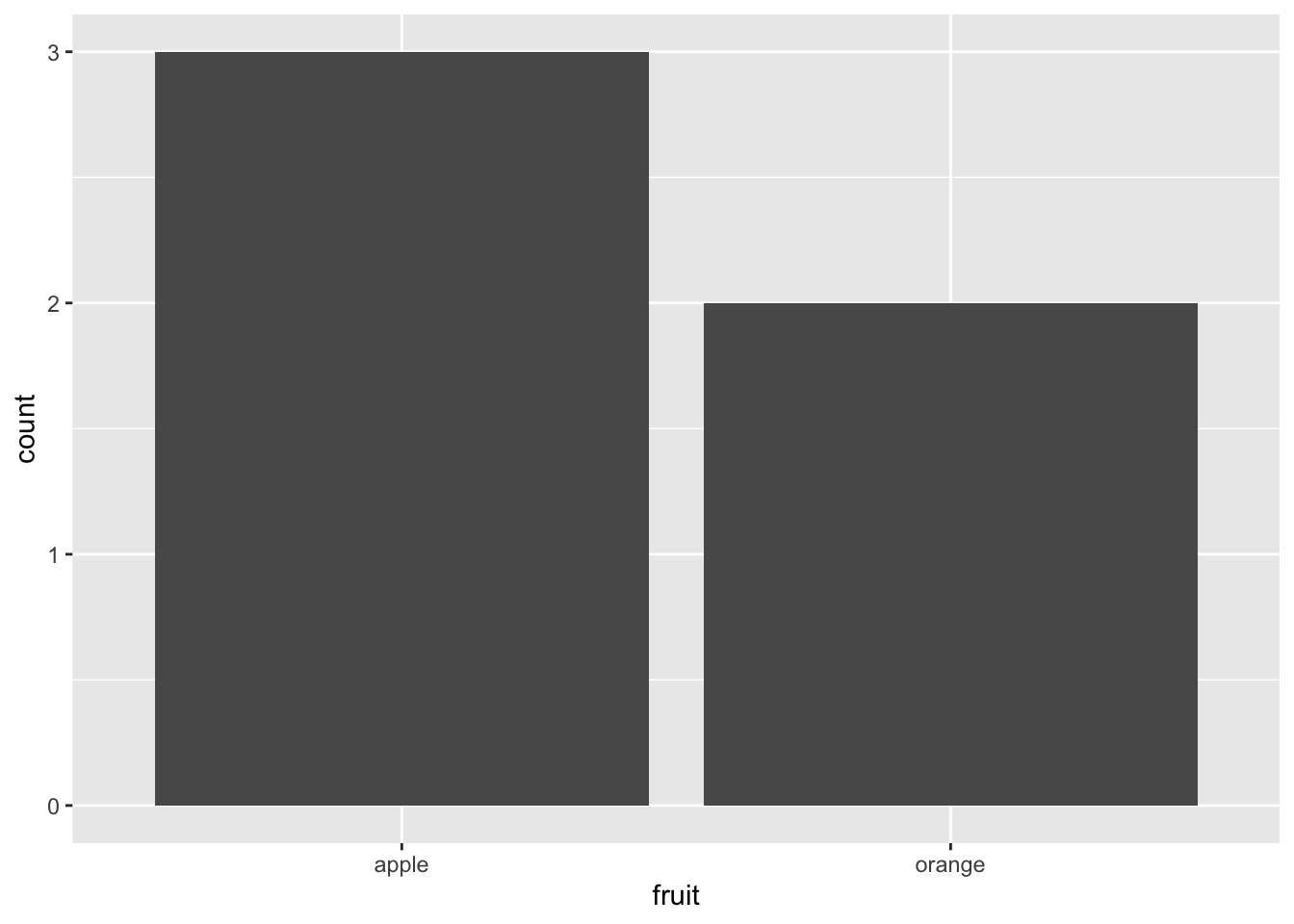
Figure 2.18: Barplot when counts are not pre-counted.
However, using the fruits_counted data frame
where the fruits have been “pre-counted,”
we once again map the fruit variable to the x-position aesthetic,
but here we also map the count variable to the y-position aesthetic,
and add a geom_col() layer instead.
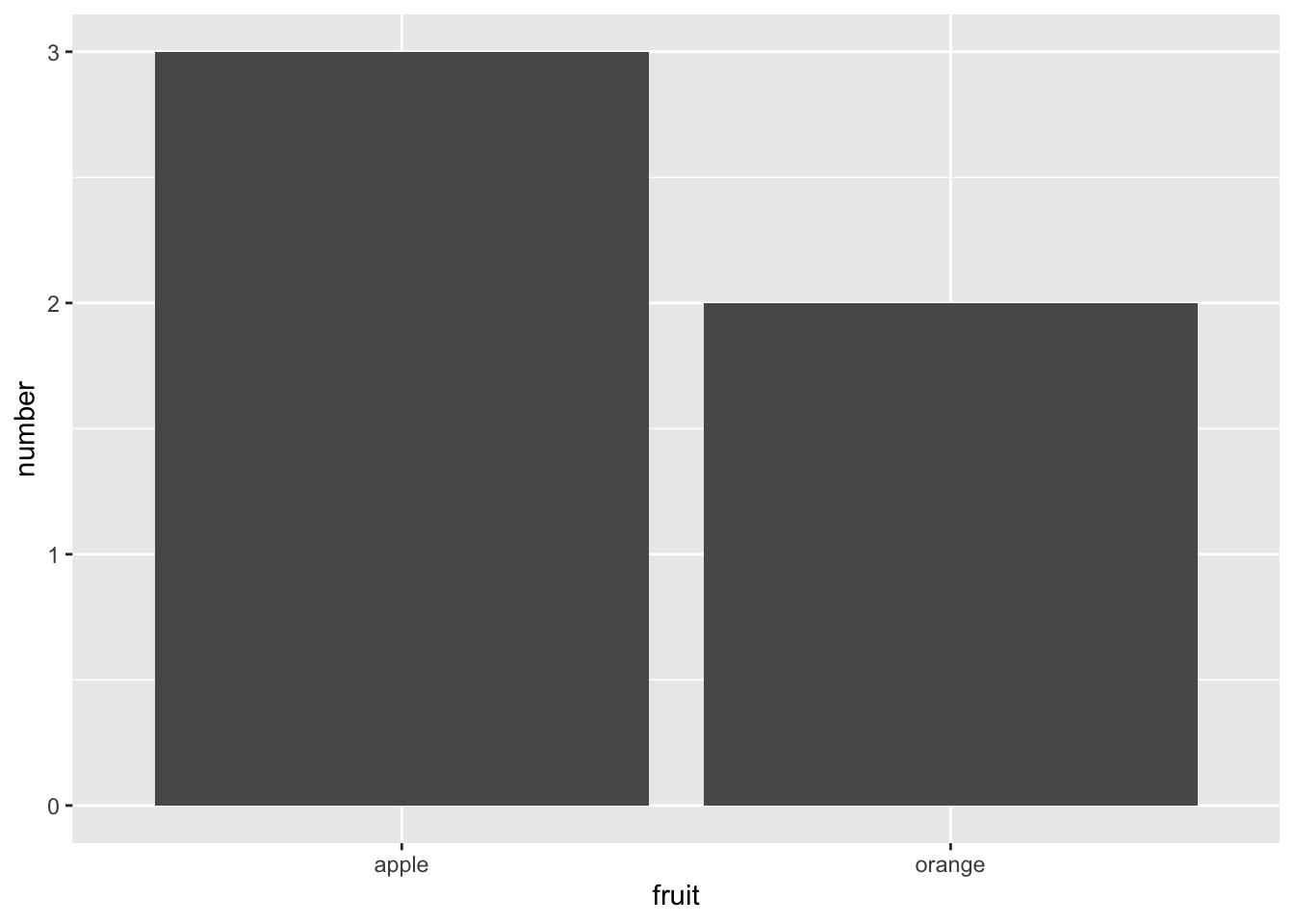
Figure 2.19: Barplot when counts are pre-counted.
Compare the barplots in Figures 2.18 and 2.19.
They are identical because they reflect counts of the same five fruits.
However, depending on how our categorical data is represented,
either “pre-counted” or not, we must add a different geom layer.
When the categorical variable whose distribution you want to visualize
- Is not pre-counted in your data frame, we use
geom_bar(). - Is pre-counted in your data frame, we use
geom_col()with the y-position aesthetic mapped to the variable that has the counts.
Let’s now go back to the df_flights data frame in the nycflights13 package
and visualize the distribution of the categorical variable carrier.
In other words, let’s visualize the number of domestic flights
out of New York City each airline company flew in 2013.
Recall from Subsection 1.4.2
when you first explored the df_flights data frame,
you saw that each row corresponds to a flight.
In other words, the df_flights data frame is more like the fruits data frame
than the fruits_counted data frame
because the flights have not been pre-counted by carrier.
Thus we should use geom_bar() instead of geom_col() to create a barplot.
Much like a geom_histogram(),
there is only one variable in the aes() aesthetic mapping:
the variable carrier gets mapped to the x-position.
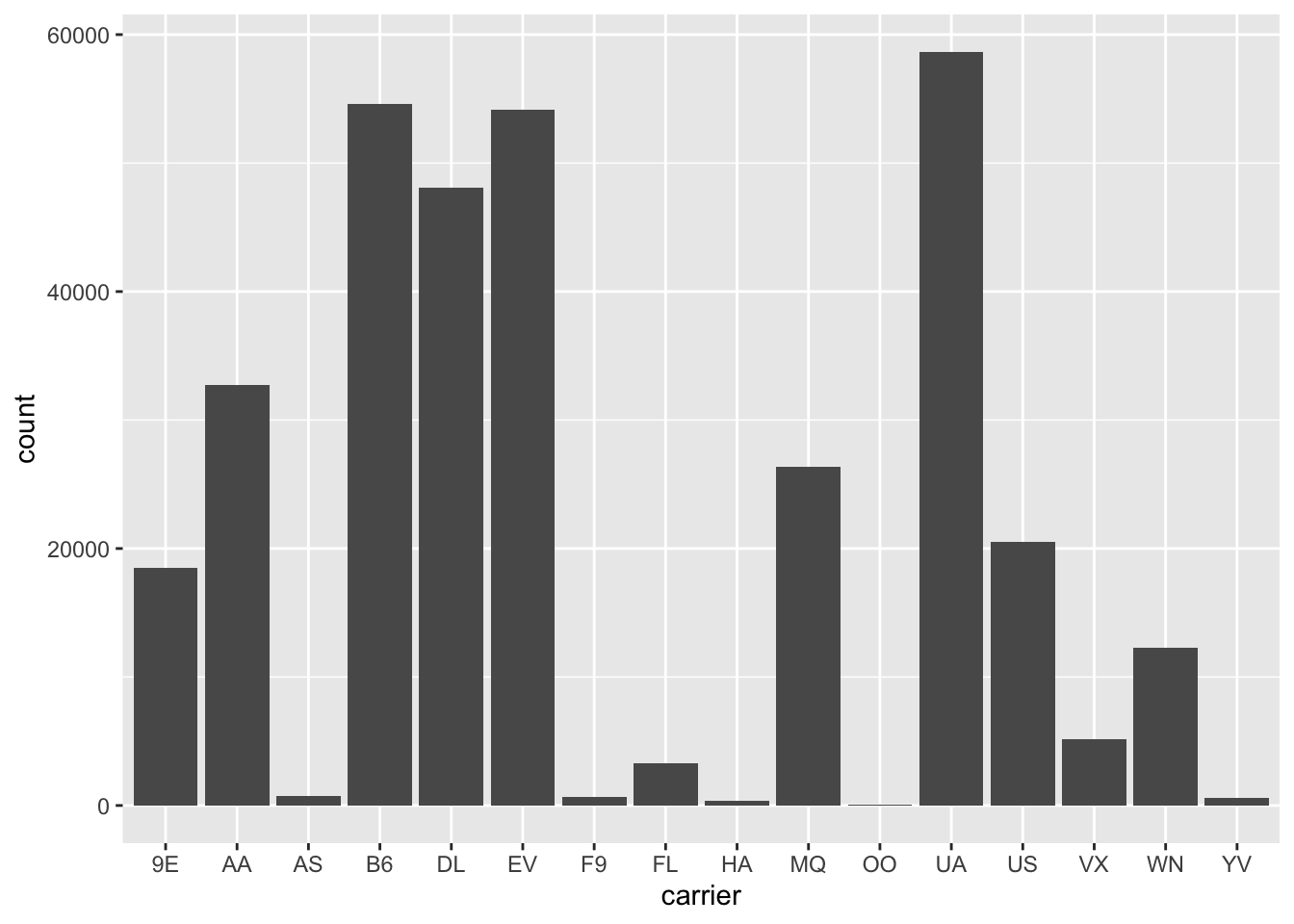
Figure 2.20: Number of flights departing NYC in 2013 by airline using geom_bar().
Observe in Figure 2.20 that United Airlines (UA),
JetBlue Airways (B6), and ExpressJet Airlines (EV)
had the most flights depart NYC in 2013.
If you don’t know which airlines correspond to which carrier codes,
then run View(nycflights13::airlines)
to see a directory of airlines.
For example, B6 is JetBlue Airways.
Alternatively, say you had a data frame where the number of flights
for each carrier was pre-counted as in Table 2.4.
| carrier | number |
|---|---|
| 9E | 18460 |
| AA | 32729 |
| AS | 714 |
| B6 | 54635 |
| DL | 48110 |
| EV | 54173 |
| F9 | 685 |
| FL | 3260 |
| HA | 342 |
| MQ | 26397 |
| OO | 32 |
| UA | 58665 |
| US | 20536 |
| VX | 5162 |
| WN | 12275 |
| YV | 601 |
In order to create a barplot visualizing the distribution
of the categorical variable carrier in this case,
we would now use geom_col() instead of geom_bar(),
with an additional y = number in the aesthetic mapping
on top of the x = carrier.
The resulting barplot would be identical to Figure 2.20.
Learning check
(LC2.22) When would a barplot be more appropriate than a histogram? Hint: consider which type of variables are being visulized.
(LC2.23) What is the difference between histograms and barplots?
(LC2.24) How many Envoy Air flights departed NYC in 2013?
(LC2.25) What was the 7th highest airline for departed flights from NYC in 2013? How could we better present the table to get this answer quickly?
2.8.2 Must avoid pie charts!
One of the most common plots used to visualize the distribution of categorical data is the pie chart. While they may seem harmless enough, pie charts actually present a problem in that humans are unable to judge angles well. As Naomi Robbins describes in her book, Creating More Effective Graphs (Robbins 2013), we overestimate angles greater than 90 degrees and we underestimate angles less than 90 degrees. In other words, it is difficult for us to determine the relative size of one piece of the pie compared to another.
Let’s examine the same data used in our previous barplot of the number of flights departing NYC by airline in Figure 2.20, but this time we will use a pie chart in Figure 2.21. Try to answer the following questions:
- How much larger is the portion of the pie for
ExpressJet Airlines (
EV) compared to US Airways (US)? - What is the third largest carrier in terms of departing flights?
- How many carriers have fewer flights than United Airlines (
UA)?
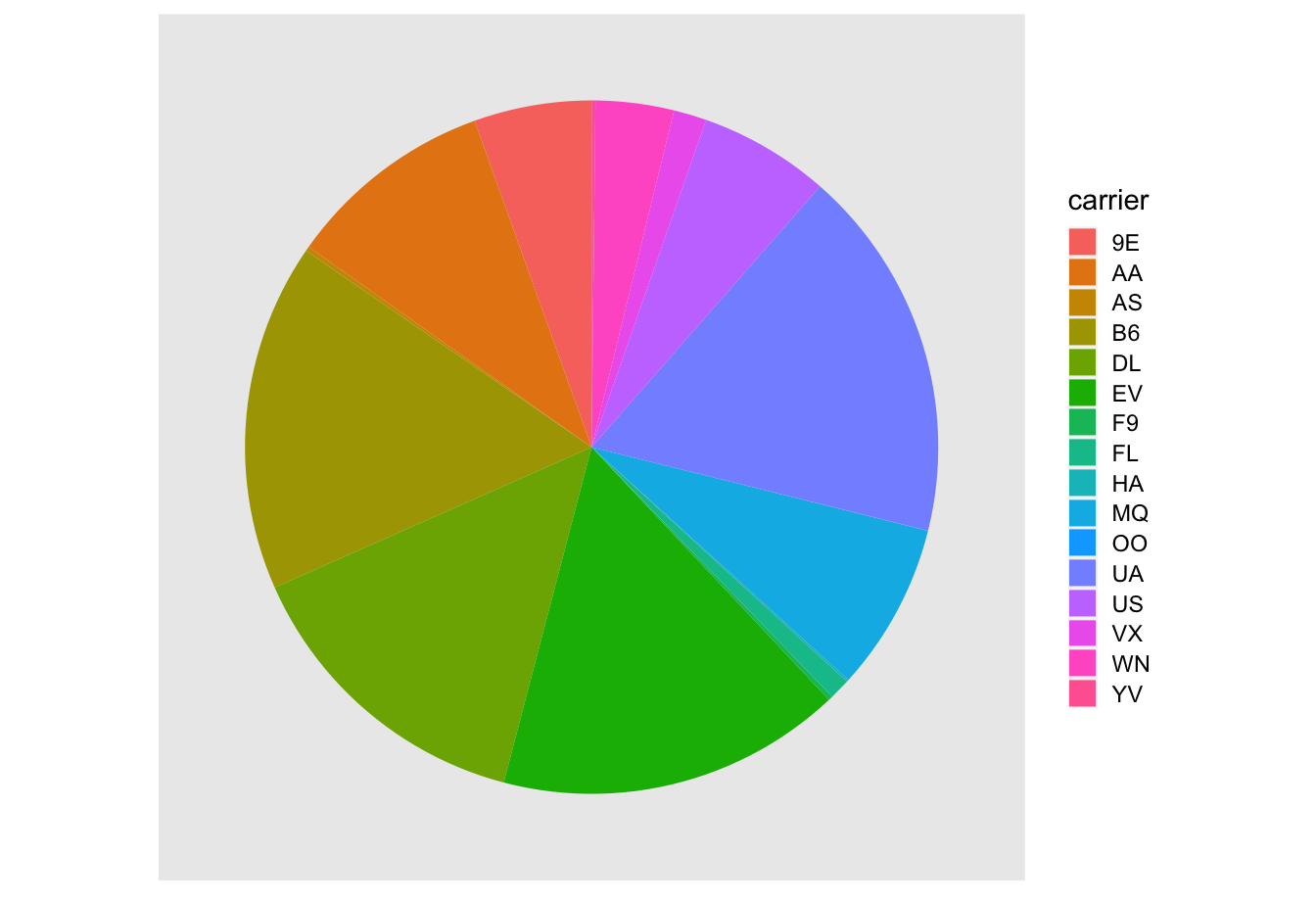
Figure 2.21: The dreaded pie chart.
While it is quite difficult to answer these questions when looking at the pie chart in Figure 2.21, we can much more easily answer these questions using the barchart in Figure 2.20. This is true since barplots present the information in a way such that comparisons between categories can be made with single horizontal lines, whereas pie charts present the information in a way such that comparisons must be made by comparing angles.
Learning check
(LC2.26) Why do you think people continue to use pie charts?
2.8.3 Two categorical variables
Barplots are a very common way to visualize the frequency
of different categories, or levels, of a single categorical variable.
Another use of barplots is to visualize the joint distribution
of two categorical variables at the same time.
Let’s examine the joint distribution of outgoing domestic flights
from NYC by carrier as well as origin.
In other words, the number of flights
for each carrier and origin combination.
For example, the number of WestJet flights from JFK,
the number of WestJet flights from LGA,
the number of WestJet flights from EWR,
the number of American Airlines flights from JFK,
and so on.
Recall the ggplot() code that created the barplot of carrier frequency
in Figure 2.20:
We can now map the additional variable origin
by adding a fill = origin inside the aes() aesthetic mapping.
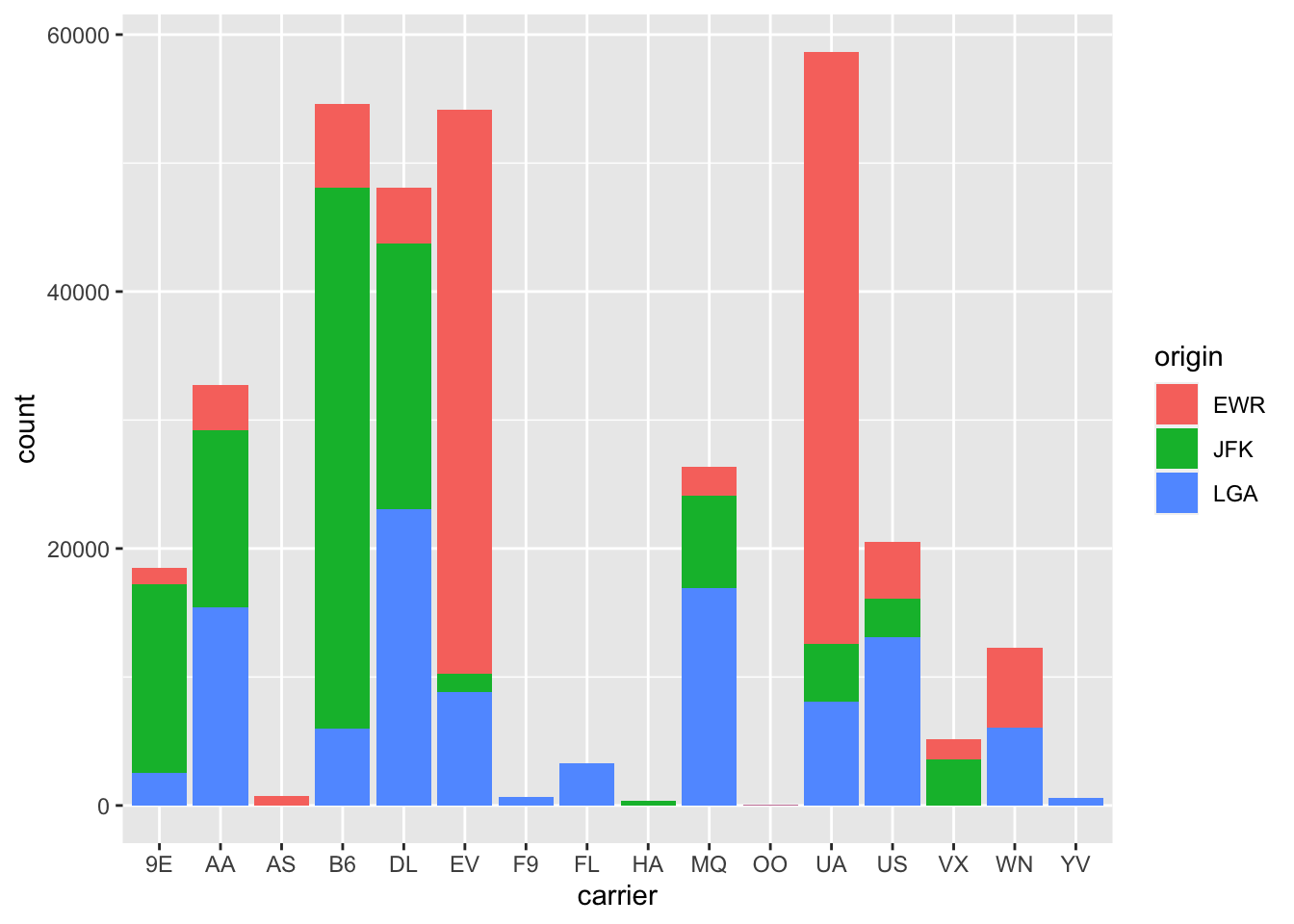
Figure 2.22: Stacked barplot of flight amount by carrier and origin.
Figure 2.22 is an example
of a stacked barplot.
While simple to make, in certain aspects it is not ideal.
For example, it is difficult to compare the heights
of the different colors between the bars,
corresponding to comparing the number of flights
from each origin airport between the carriers.
Before we continue, let’s address some common points of
confusion among new R users.
First, the fill aesthetic corresponds to the color used to fill the bars,
while the color aesthetic corresponds to the color of the outline of the bars.
This is identical to how we added color to our histogram
in Subsection 2.5.1:
we set the outline of the bars to white by setting color = "white"
and the colors of the bars to blue steel by setting fill = "steelblue".
Observe in Figure 2.23
that mapping origin to color and not fill
yields grey bars with different colored outlines.
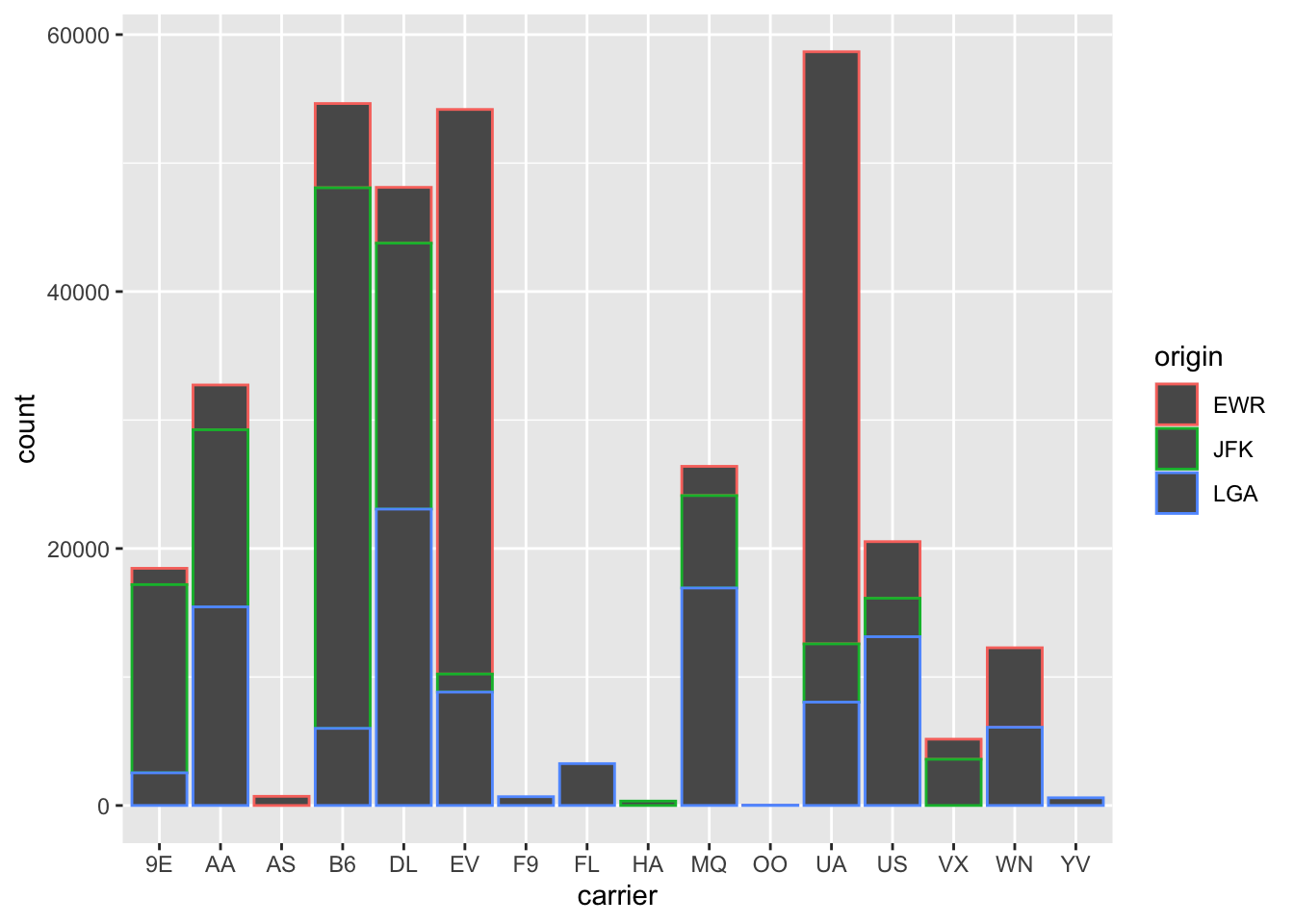
Figure 2.23: Stacked barplot with color aesthetic used instead of fill.
Second, note that fill is another aesthetic mapping
much like x-position;
thus we were careful to include it
within the parentheses of the aes() mapping.
The following code, where the fill aesthetic is specified
outside the aes() mapping will yield an error.
This is a fairly common error that new ggplot users make:
# Try on your own. Will yield an error.
gg$ggplot(data = df_flights,
mapping = gg$aes(x = carrier), fill = origin) +
gg$geom_bar()An alternative to stacked barplots are
side-by-side barplots,
also known as dodged barplots,
as seen in Figure 2.24.
The code to create a side-by-side barplot
is identical to the code to create a stacked barplot,
but with a position = "dodge" argument
added to geom_bar().
In other words, we are overriding the default barplot type,
which is a stacked barplot,
and specifying it to be a side-by-side barplot instead.
gg$ggplot(data = df_flights,
mapping = gg$aes(x = carrier, fill = origin)) +
gg$geom_bar(position = "dodge")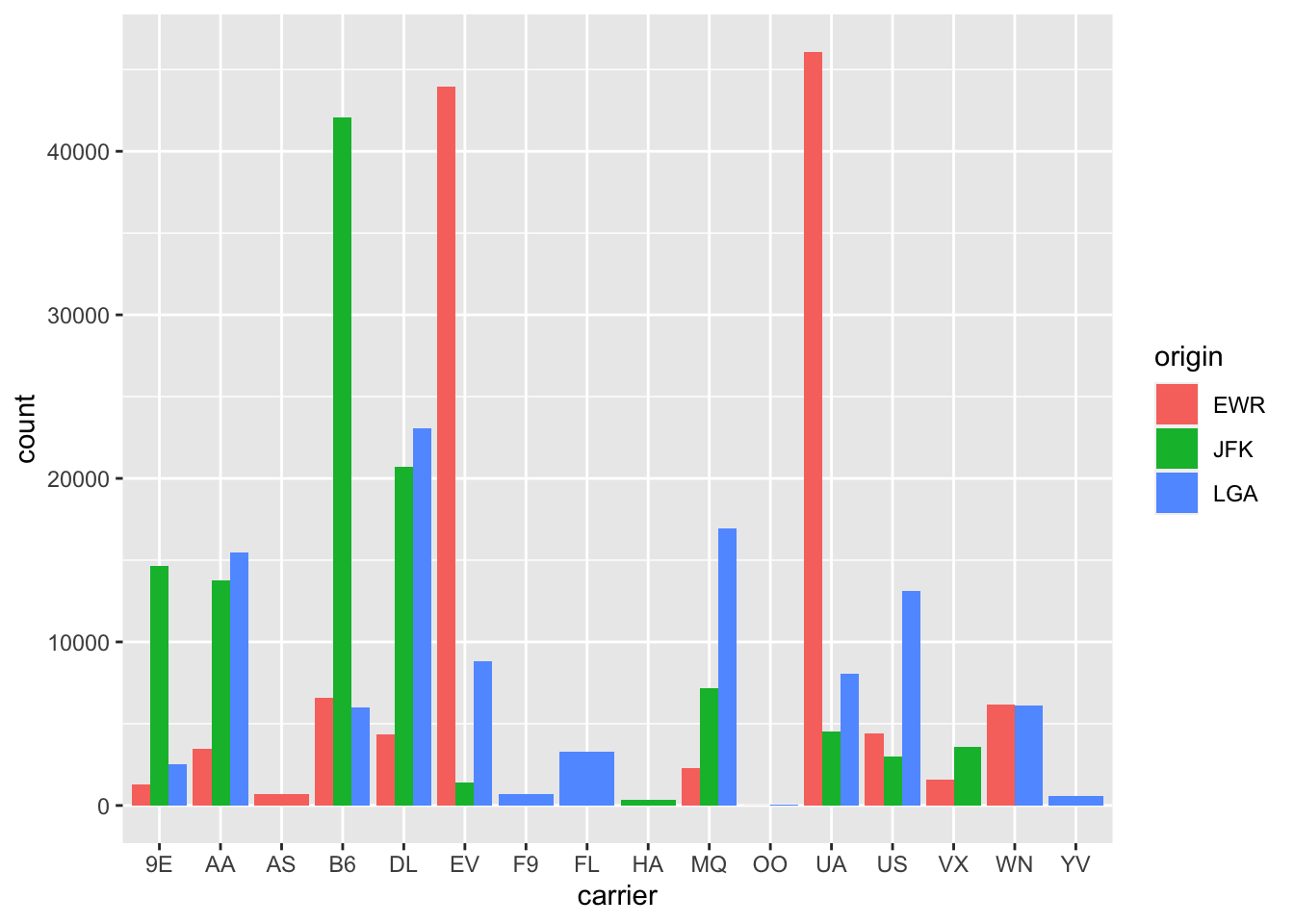
Figure 2.24: Side-by-side barplot comparing number of flights by carrier and origin.
Note the width of the bars for AS, F9, FL, HA and YV
is different than the others.
We can make one tweak to the position argument
to get them to be the same size in terms of width
as the other bars by using the more robust position_dodge() function.
gg$ggplot(data = df_flights,
mapping = gg$aes(x = carrier, fill = origin)) +
gg$geom_bar(position = gg$position_dodge(preserve = "single"))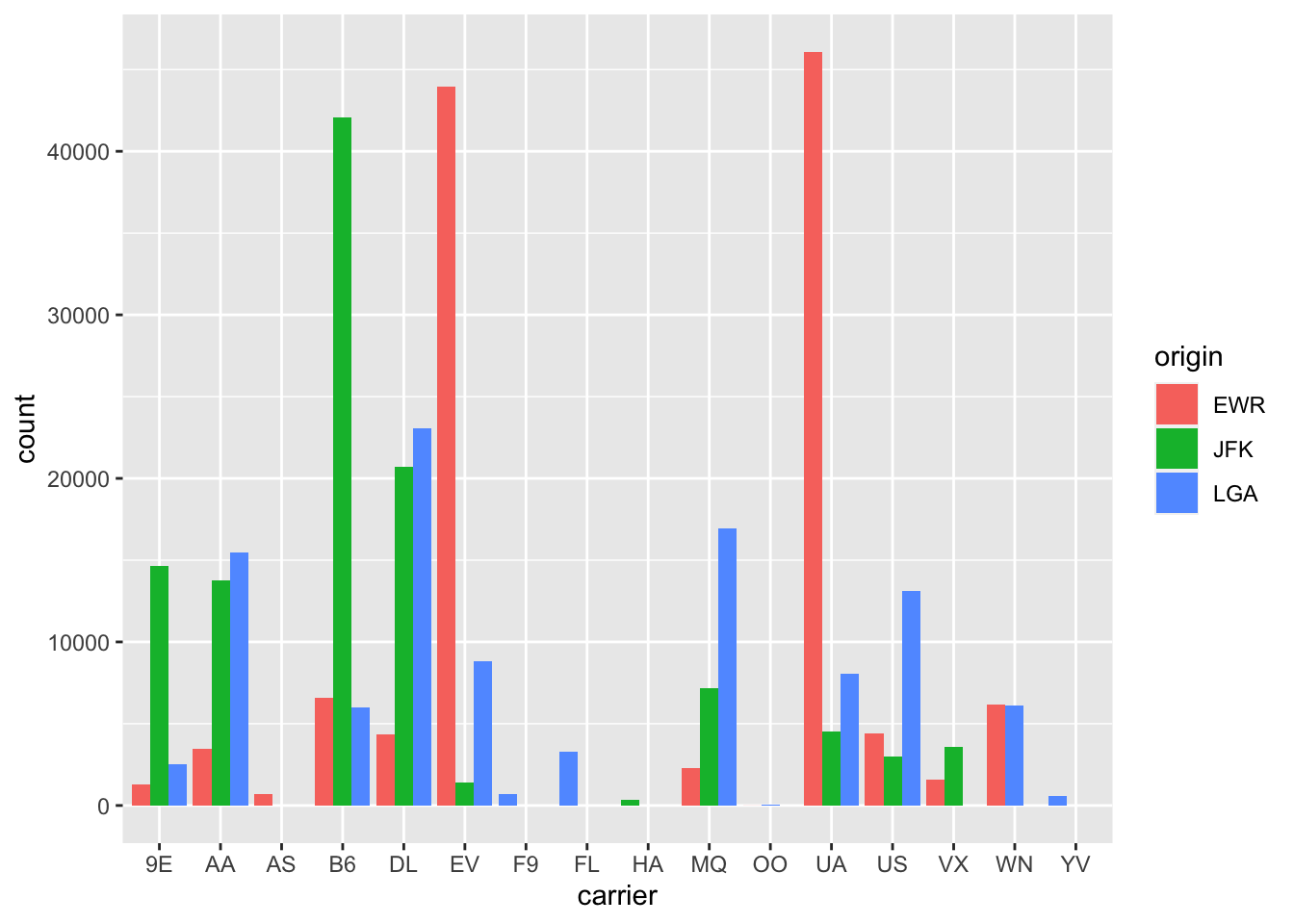
Figure 2.25: Side-by-side barplot comparing number of flights by carrier and origin (with formatting tweak).
Lastly, another type of barplot is a faceted barplot.
Recall in Section 2.6 we visualized the distribution
of hourly temperatures at the 3 NYC airports split by month using facets.
We apply the same principle to our barplot
visualizing the frequency of carrier split by origin:
instead of mapping origin to fill
we include it as the variable to create small multiples of the plot
across the levels of origin.
gg$ggplot(data = df_flights,
mapping = gg$aes(x = carrier)) +
gg$geom_bar() +
gg$facet_wrap(~ origin, ncol = 1)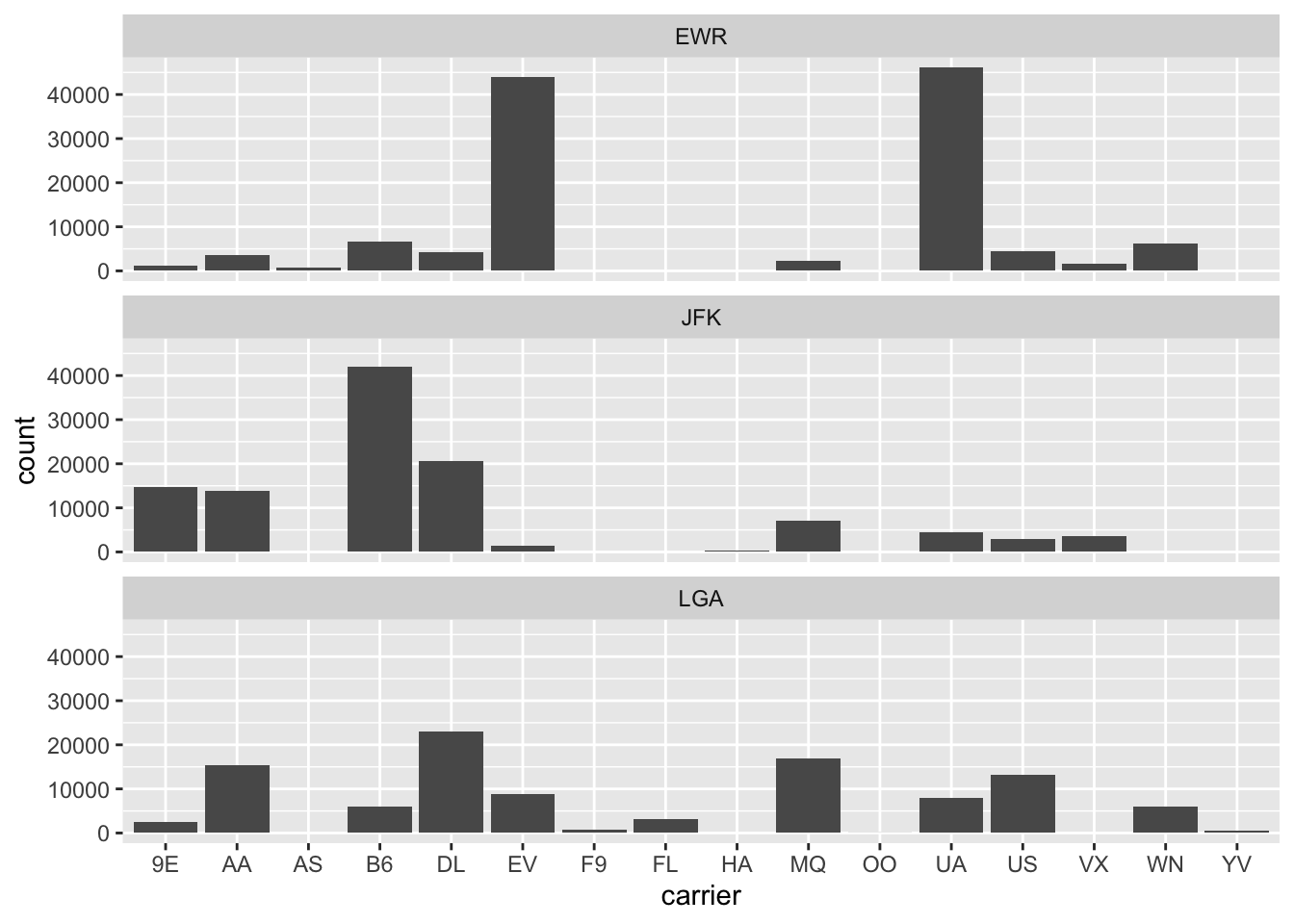
Figure 2.26: Faceted barplot comparing the number of flights by carrier and origin.
Learning check
(LC2.27) What kinds of questions are not easily answered by looking at Figure 2.22?
(LC2.28) What can you say, if anything, about the relationship between airline and airport in NYC in 2013 in regards to the number of departing flights?
(LC2.29) Why might the side-by-side barplot be preferable to a stacked barplot in this case?
(LC2.30) What are the disadvantages of using a dodged barplot, in general?
(LC2.31) Why is the faceted barplot preferred to the side-by-side and stacked barplots in this case?
(LC2.32) What information about the different carriers at different airports is more easily seen in the faceted barplot?
2.8.4 Summary
Barplots are a common way of displaying the distribution
of a categorical variable,
or in other words the frequency with which the different categories
(also called levels) occur.
They are easy to understand and make it easy to make comparisons across levels.
Furthermore, when trying to visualize the relationship
of two categorical variables,
you have many options: stacked barplots, side-by-side barplots,
and faceted barplots.
Depending on what aspect of the relationship you are trying to emphasize,
you will need to make a choice
between these three types of barplots and own that choice.
2.9 Conclusion
2.9.1 Summary table
Let’s recap all five of the five named graphs (5NG)
in Table 2.5
summarizing their differences.
Using these 5NG,
you’ll be able to visualize the distributions and relationships
of variables contained in a wide array of datasets.
This will be even more the case as we start to map more variables
to more of each geometric object’s aesthetic attribute options,
further unlocking the awesome power of the ggplot2 package.
| Named graph | Shows | Geometric object | Notes | |
|---|---|---|---|---|
| 1 | Scatterplot | Relationship between 2 continuous variables |
geom_point()
|
NA |
| 2 | Linegraph | Relationship between 2 continuous variables |
geom_line()
|
Used when there is a sequential order to x-variable, e.g., time |
| 3 | Histogram | Distribution of 1 continuous variable |
geom_histogram()
|
Facetted histograms show the distribution of 1 continuous variable split by the values of another variable |
| 4 | Boxplot | Distribution of 1 continuous variable split by the values of another variable |
geom_boxplot()
|
NA |
| 5 | Barplot | Distribution of 1 categorical variable |
geom_bar() when counts are not pre-counted, geom_col() when counts are pre-counted
|
Stacked, side-by-side, and faceted barplots show the joint distribution of 2 categorical variables |
2.9.2 Function argument specification
Let’s go over some important points about specifying the arguments (i.e., inputs) to functions. Run the following two segments of code:
# Segment 1:
gg$ggplot(data = df_flights,
mapping = gg$aes(x = carrier)) +
gg$geom_bar()
# Segment 2:
gg$ggplot(df_flights,
gg$aes(x = carrier)) +
gg$geom_bar()You’ll notice that both code segments create the same barplot,
even though in the second segment
we omitted the data = and mapping = code argument names.
This is because the ggplot() function by default assumes that
the data argument comes first
and the mapping argument comes second.
As long as you specify the data frame in question first
and the aes() mapping second,
you can omit the explicit statement of the argument names
data = and mapping =.
Going forward for the rest of this book,
all ggplot() code will be like the second segment:
with the data = and mapping = explicit naming of the argument
omitted with the default ordering of arguments respected.
We’ll do this for brevity’s sake;
it’s common to see this style when reviewing other R users’ code.
2.10 Additional resources
2.10.1 factor() in boxplot
In section 2.7.1, we learned how to make side-by-side boxplots with the following code:
gg$ggplot(data = df_weather,
mapping = gg$aes(x = factor(month, ordered=T), y = temp_c)) +
gg$geom_boxplot()Structure-wise, this code resembles what we used to create scatterplots and linegraphs, which I reproduce below for you to compare:
# scatterplot
gg$ggplot(data = df_alaska_flights,
mapping = gg$aes(x = dep_delay, y = arr_delay)) +
gg$geom_point() + # linegraph
gg$ggplot(data = early_january_weather,
mapping = gg$aes(x = time_hour, y = temp_c)) +
gg$geom_line()There is one distinguished feature in the boxplot code though:
mapping = gg$aes(x = factor(month, ordered=T)) ...Instead of x = month, we used x = factor(month, ordered=T).
Naturally, you will ask what we can achieve with factor()?
And why do we need to apply factor() on the month variable
for the boxplot?
factor() is a base R function.
It is typically applied to variables that are inherently categorical in nature,
such as employment status, or days of a week.
These variables share a common feature:
they all have a countable number of possible values,
rather than having a countless number of possible values.
Take days of a week versus the height of a person for example.
There are 7 possible values for the former: Monday, Tuesday,
Wednesday, Thursday, Friday, Saturday and Sunday.
In contrast, there is an infinite number of possible values
for a person’s height:
1.6 m, 1.61 m, 1.611 m, 1.6111 m, … You get the idea.
Variables that have a finite number of possible values are often called
categorical or discrete variables,
whereas those that have an infinite number of possible values
are called continuous variables.
Note the italicized word possible in the previous paragraph.
When judging whether a variable is categorical or continuous,
it is important to think what its possible values are,
rather than what its actual values are in a sample.
A height variable may only have a finite number of actual values
in a sample of 10 people: 1.58 m, 1.63 m, 1.79 m, 1.67 m, 1.72 m, 1.65 m,
1.74 m, 1.82 m, 1.79 m, 1.81 m.
However, as we have illustrated before,
the possibilities are infinite.
Therefore, height would be considered a continuous variable.
Let’s bring our attention back to the boxplot.
x = factor(month, ordered=T)
In this example,
month has exactly 12 possible values: “1” through “12.”
By applying factor() to the variable month,
we are giving R explicit instruction to treat month as a categorical variable,
with 12 levels.
In addition, by setting ordered to T (short for TRUE),
we are practically telling R to sort the 12 levels of month,
with 1 being the first and 12 the last,
matching the natural order of twelve months in a calendar year.
What happens if you do not apply factor() to month,
and just use month as is for the boxplot?
Let’s try it!
gg$ggplot(data = df_weather,
mapping = gg$aes(x = month, y = temp_c)) +
gg$geom_boxplot() +
gg$labs(y = "Temperature (Celsius)")Warning: Continuous x aesthetic -- did you forget aes(group=...)?Warning: Removed 1 rows containing non-finite values (stat_boxplot).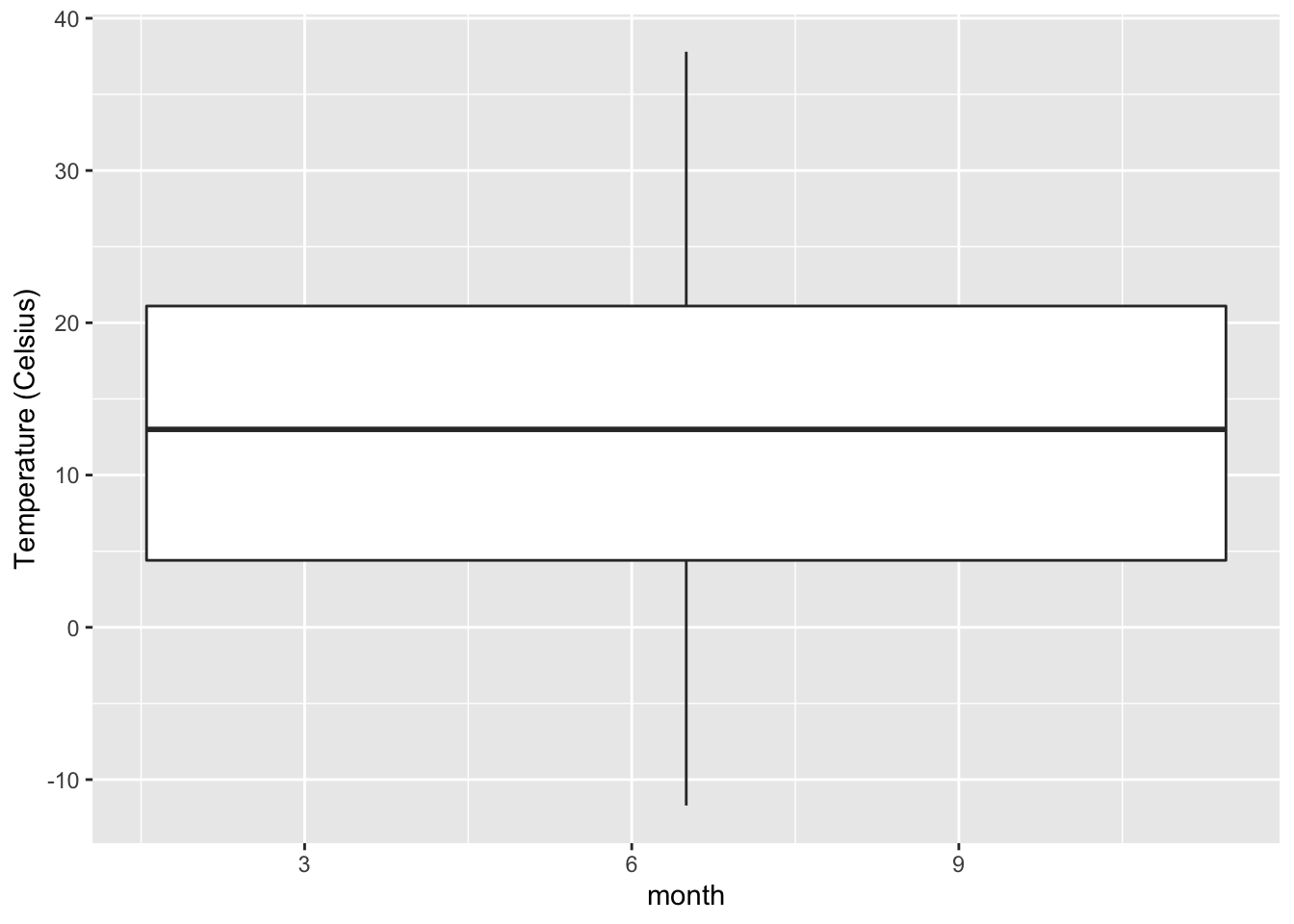
Figure 2.27: Invalid boxplot specification.
Observe in Figure 2.27 that this plot
does not look like Figure 2.17 at all.
Unlike Figure 2.17,
it does not provide information about temperature separated by month.
The first warning message clues us in as to why.
It is telling us that we have a “continuous” variable on the x-axis.
But you may protest:
is it not obvious that the month variable is a categorical one?
After all, there are only “1” through “12,” tweleve actual values
in the data df_weather.
Yes, as an adult English speaker,
it is very obivous that month is a categorical variable,
evidenced by its values only spanning the twelve integers 1 to 12.
However, R is not an adult English speaker.
If anything, it is more like an utterly stubborn polylingual rule stickler.
It demands explicit instruction from us, the superior human users,
on how to interpret a variable.
Until we explicitly tell R that month is a categorical variable
with ordered levels 1 through 12,
it will treat month as a continuous one,
with possible values of 1, 2, 3, …, 10, 11, 12, 13, 14, …
So we have seen what happens if we omit the factor(..) in the boxplot.
ggplot’s boxplots require a categorical variable to be mapped
to the x-position aesthetic.
When the requirement is not met,
ggplot throws out the unsatisfying month,
gives a warning,
and tries to only work with the y-axis, temp_c,
which results in Figure 2.27.
The second warning message is equivalent to the warning message we have seen
when plotting a histogram of hourly temperatures
in Figure 2.10:
that one of the rows has missing value NA in temp_c and/or month.
To confirm:
| origin | year | month | day | hour | temp | dewp | humid | wind_dir | wind_speed | wind_gust | precip | pressure | visib | time_hour | temp_c |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EWR | 2013 | 8 | 22 | 9 | NA | NA | NA | 320 | 12.65858 | NA | 0.13 | NA | 7 | 2013-08-22 09:00:00 | NA |
Sure enough, this row has a missing value in temp_c.
(Place your mouse cursor inside the table and scroll right
to see more columns.)
Learning check
(LC2.33)
What happens when you completely drop the x axis in the boxplot?
(LC2.34)
We looked at the distribution of the continuous variable temp_c
split by the variable month that we converted
using the factor() function in order to make a side-by-side boxplot.
Why would a boxplot of temp_c split by the continuous variable pressure
similarly converted to a categorical variable
using the factor() not be informative?
2.10.2 Common problems
This section is from https://r4ds.had.co.nz/data-visualisation.html#common-problems
As you start to run R code, you’re likely to run into problems. Don’t worry — it happens to everyone. I have been writing R code for years, and every day I still write code that doesn’t work!
Start by carefully comparing the code that you’re running
to the code in the book.
R is extremely picky, and a misplaced character can make all the difference.
Make sure that every ( is matched with a )
and every " is paired with another ".
Sometimes you’ll run the code and nothing happens.
Check the left-hand of your console: if it’s a +,
it means that R doesn’t think you’ve typed a complete expression
and it’s waiting for you to finish it.
In this case, it’s usually easy to start from scratch again
by pressing ESCAPE to abort processing the current command.
One common problem when creating ggplot2 graphics
is to put the + in the wrong place:
it has to come at the end of the line, not the start.
In other words, make sure you haven’t accidentally written code like this:
# Try by yourself, will encounter an error
ggplot2::ggplot(data = mpg)
+ ggplot::geom_point(
mapping = ggplot2::aes(x = displ, y = hwy))If you’re still stuck, try the help (see section 1.4.4). Don’t worry if the help doesn’t seem that helpful - instead, skip down to the examples and look for code that matches what you’re trying to do.
If that doesn’t help, carefully read the error message. Sometimes the answer will be buried there! But when you’re new to R, the answer might be in the error message but you don’t yet know how to understand it. Another great tool is Google: try googling the error message, as it’s likely someone else has had the same problem, and has gotten help online.
2.10.3 What’s to come
Recall in Figure 2.2 in Section 2.3
we visualized the relationship between departure delay and arrival delay
for Alaska Airlines flights.
This necessitated paring down the df_flights data frame
to a new data frame alaska_flights
consisting of only carrier == AS flights first:
alaska_flights <- df_flights %>%
dp$filter(carrier == "AS")
gg$ggplot(data = alaska_flights,
mapping = gg$aes(x = dep_delay, y = arr_delay)) +
gg$geom_point()Furthermore recall in Figure 2.7 in Section 2.4
we visualized hourly temperature recordings at Newark airport
only for the first 15 days of January 2013.
This necessitated paring down the df_weather data frame
to a new data frame early_january_weather
consisting of hourly temperature recordings only for origin == "EWR",
month == 1, and day less than or equal to 15 first:
early_january_weather <- df_weather %>%
dp$filter(origin == "EWR" & month == 1 & day <= 15)
gg$ggplot(data = early_january_weather,
mapping = gg$aes(x = time_hour, y = temp_c)) +
gg$geom_line()These two code segments were a preview of Chapter 3
on data wrangling using the dplyr package.
Data wrangling is the process of transforming
and modifying existing data with the intent of making it more appropriate
for analysis purposes.
For example, these two code segments used the filter() function
to create new data frames (alaska_flights and early_january_weather)
by choosing only a subset of rows of existing data frames
(flights and weather).
In the next chapter, we’ll formally introduce the filter()
and other data wrangling functions
as well as the pipe operator %>%
which allows you to combine multiple data wrangling actions
into a single sequential chain of actions.
On to Chapter 3 on data wrangling!