Appendix C Trouble shooting and tips
In this section, I compiled a list of questions that I received from you, as well as questions that I anticipate you may have at some point regarding R & RStudio. Each question is accompanied by an answer. The list will be updated regularly.
C.1 Questions and answers
C.1.1 When do I install?
Do I need to install
dplyrevery time I re-open my RStudio or it is now available all the time in my current sandbox?
Response:
Installing an R package is similar to installing an app on your phone. You only need to install it once, and maybe reinstall it later for update. However, to be able to use an app on your phone, not only do you have to have it installed (once), you also need to open the app each time you turn on your phone. The action of opening an app, for an R package, is equivalent to importing the package.
C.1.2 When do I import?
I believe I have run
import::from(magrittr, "%>%")yesterday so I wasn’t aware that I should run again every time I reopen my Rstudio
Response:
Just as you need to open an app on your phone each time you want to use it,
you need to import each time you want to use a function.
You may import only the function you need from a package, such as %>%.
You may also import everything from a package,
such as gg <- import::from(ggplot2, .all=T, .into={new.env()}).
C.1.3 Which one should I install first?
I am confused whether I should use
install.packages("ggplot2")as a first line of my code orinstall.packages("import").
Response:
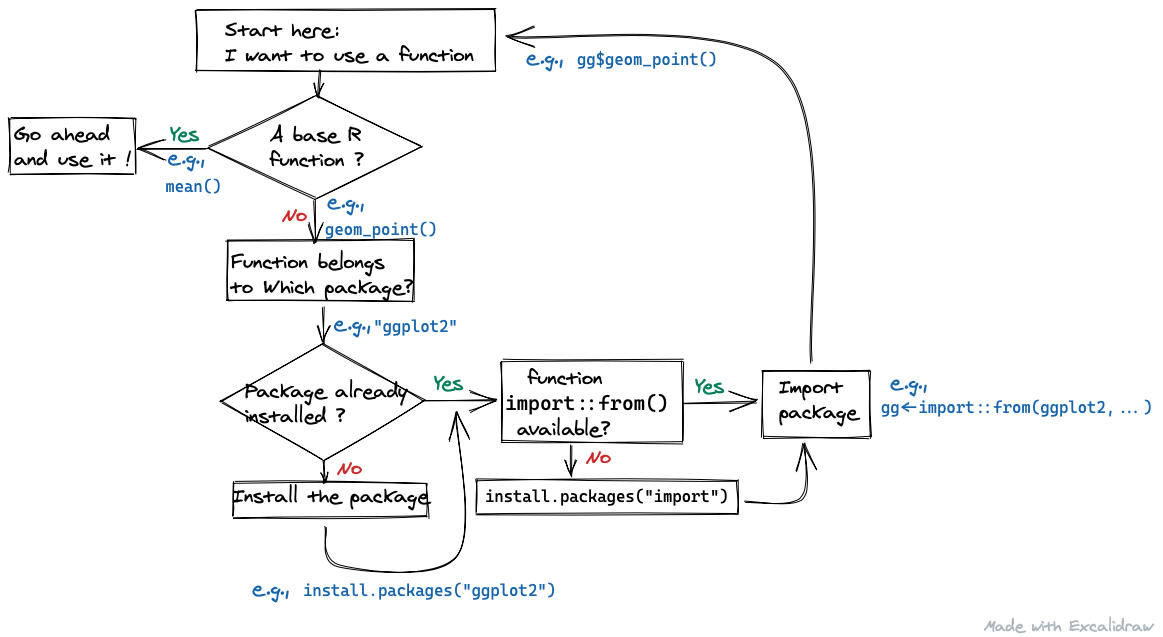
Figure C.1: When do I need to install?
C.1.4 library()
I’m confused with the
glimpse()function: when I use it in the online module on Datacamp, the commandglimpse(any_data_frame)works. But when I try it in R-studio with the available data set, it doesn’t work, but it works when I useglimpsefunction with$sign (see the screenshot).

Figure C.2: To attach or not to attach?
Response:
glimpse() is a function in the package dplyr.
As a general rule, R will complain if you call a function without
first specifying which package this function is from,
barring those functions that are part of base R.
Consider the example of sending a letter via Canada post.
Your letter will be rejected if you have written
“123 Hell Ave.” on the evelope without the city name,
even though there might be only one “123 Hell Ave.” in the entire world.
To the postman,
there could be hundreds of “123 Hell Ave.” in the world,
and they will not do the guess work for you;
which one do you want to send the letter to?
Similarly, when you try to call a function without giving the package name,
R will stop you with an error message:
could not find function xxx [because you did not provide the complete address].
One way to be specific is to always prepend the package name
when you try to call a function, such as dplyr::glimpse().
However, typing the package name plus the double colon ::
each time when we need to use a function can become cumbersome.
As a workaround, we use import::from() to create a shortcut.
For example, instead of having to type dplyr::glimpse() each time,
we just type dp$glimpse() for short.
Revisit section 1.3.3 if you want to brush up your memory
about this topic.
Another way, which is adopted by Datacamp,
is to execute library(package) before using any function from the package.
For example, once library(dplyr) has been executed,
you can call any function from dplyr directly, without having to
explicitly reference the package in the rest of a script.
On Datacamp, library(dplyr) has been executed behind the scene,
on your behalf.
C.1.5 The pipe operator %>%
I am receiving this error, and I don’t know what to do, or what does it means
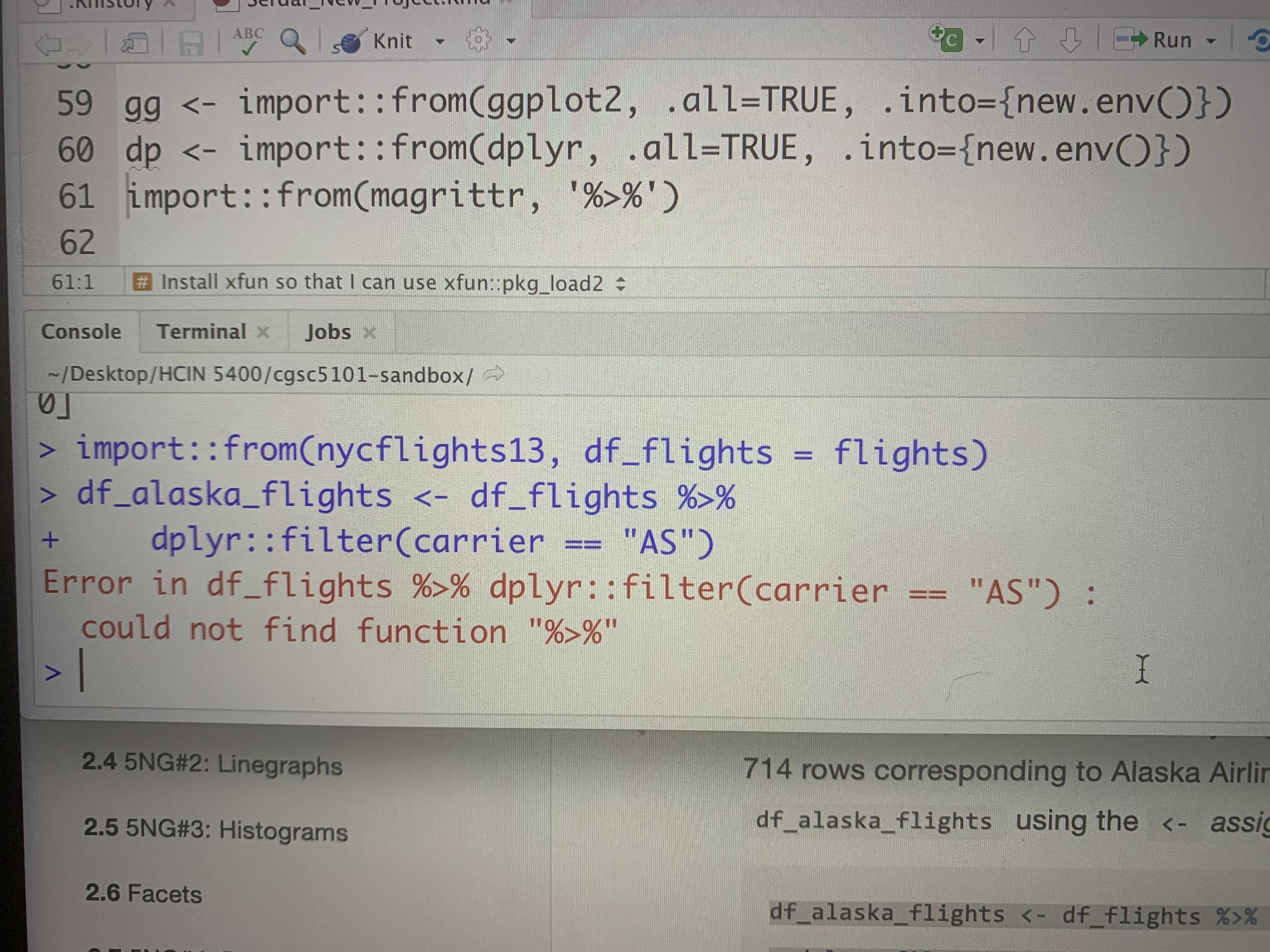
Figure C.3: could not find function ‘%>%’
Response:
could not find function xxx is a very common error.
It means that R does not know from which package to retrieve this function.
You need to first import this function from its package
before you can use it.
In this case, you need to first run import::from(magrittr, "%>%").
It may not be immediately obvious that %>% is a function.
filter() is a function; mean() is a function.
But %>%?
Yes, %>% may look different than most other functions,
because it consists of symbols, not letters.
Nevertheless, it is a function, and is often referred as the “pipe operator.”
Consider the plus sign +. It is made of a symbol, not letter.
It is an operator that operates on two numbers.
In other words, + is a function that lets us add two numbers together.
In essence, it is no different than sum(),
just in a different form.
In practice, we place two addends on both sides of +, as in 1 + 1.
Similarly, a pipe operator connects two “operands,”
such as df_flights %>% glimpse().
we need magrittr for
%>%because ggplot doesn’t provide this function?
Response:
That’s right.
Although it is common for two functions from different packages
to have the same name,
they rarely have identical funtionalities.
In the case of the pipe operator %>%,
it is only provided by the package magrittr.
C.1.6 object ‘xx’ not found
I got this error. What should I do?
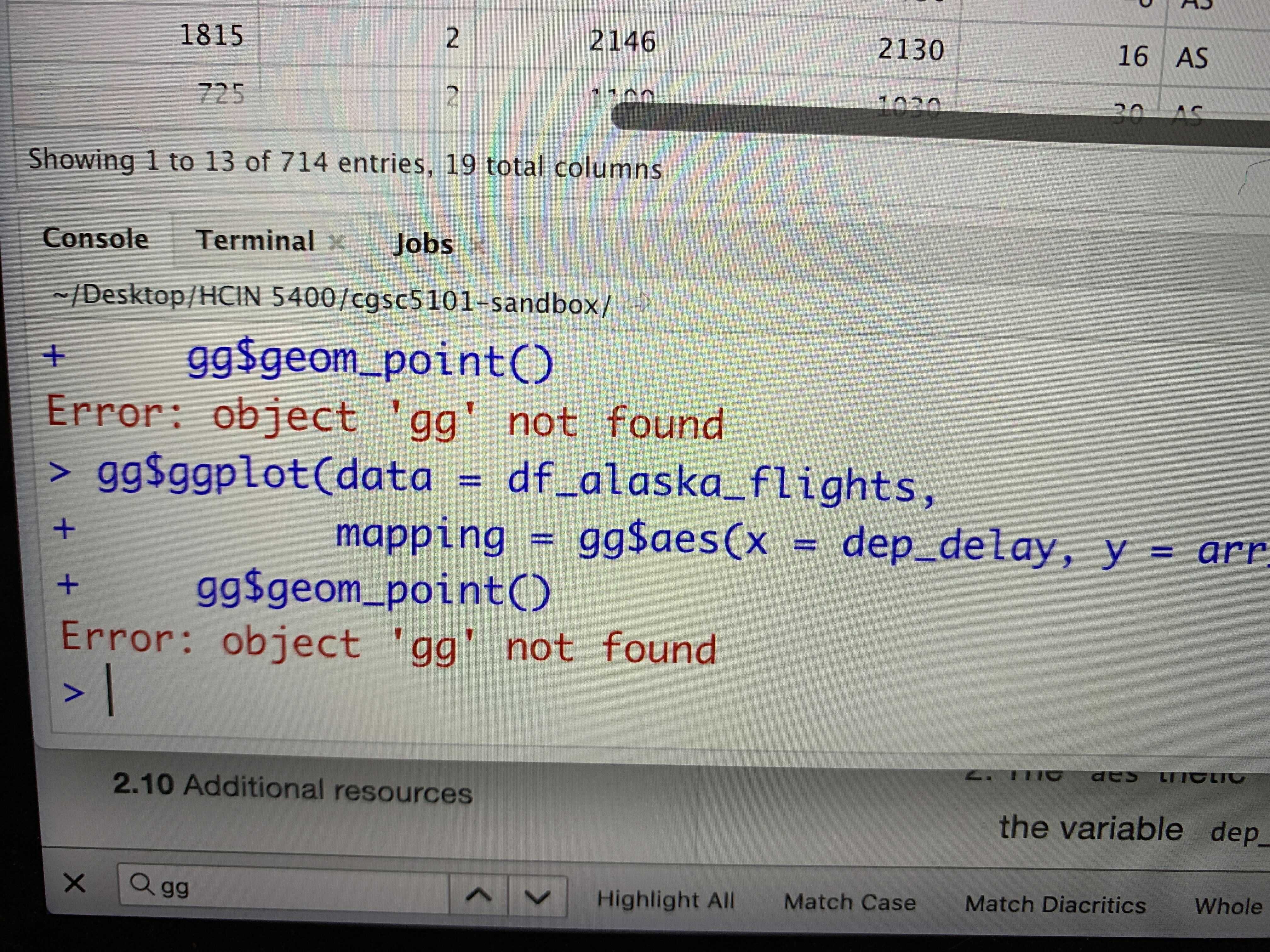
Figure C.4: object ‘gg’ not found
Response:
This error happens when you are referring to an object that has not been defined;
in this case, the object ‘gg.’
In this course, we have been using ‘gg’ as the object that
contains all the functions from package ggplot2,
as in gg <- import::from(ggplot2, .all=T, .into={new.env()}).
You may say “but I have executed / defined it yesterday.”
However, R forgets what you have executed / defined yesterday
the moment you close it.
One may consider this as inconvenient.
However, this is the beauty of all script-based software.
As the user, you are forced to record in a script
(an .R file or an .Rmd file)
what is required to reproduce your results,
be it a user-defined object or a to-be-imported package.
Only by doing so can you ensure that, when your colleague receives
your script, they can generate the same results as you did.
C.1.7 import::from()
Why do we use import::from differently in the next three examples?
Response:
In the first line, I only want to import one function, %>%,
from the package magrittr.
In the second one, I only want to import one dataset, flights,
from the package nycflights13.
I could have written
import::from(nycflights13, flights),
which would also import the dataset.
However, by including df_flights =,
I accomplish two goals simultaneously:
import the data set, and rename it as df_flights.
In the third one, I want to import everything from the package ggplot2.
I could have written
import::from(ggplot2, .all=TRUE).
However, but including gg <- and .into={new.env()},
I designate gg, an arbitrarily chosen name, as the name for a box (an object)
that would contain all the functions from ggplot2,
Doing so isolates functions from ggplot2 from base R functions
as well as funtions from other packages I may also import.
C.1.8 Unmatched parenthesis
I run the code below and I get an error: “Error: Cannot add ggproto objects together. Did you forget to add this object to a ggplot object?” How can I solve this issue?


Figure C.5: Unmatched parenthesis
Response:
There is an extra bracket after y = temp_c,
indicated by the red squiggle underneath.
A common error in R is forgetting or neglecting to finish a call
to a function with a closing ).
An example of this follows:
If you try to run the line above, R will complain:
Error in parse(text = x, srcfile = src) :
<text>:2:0: unexpected end of input
1: mean(x = c(1, 5, 10, 52)
^
Calls: <Anonymous> ... evaluate -> parse_all -> parse_all.character -> parse
Execution halted
Exited with status 1.Closing the parenthesis at the end of your call to mean
will stop the complain:
C.1.9 less than or equal to
I noticed that the order of the operators for writing less than or equal to (
<=) is important in R. The incorrect form (=<) is not recognized by R.
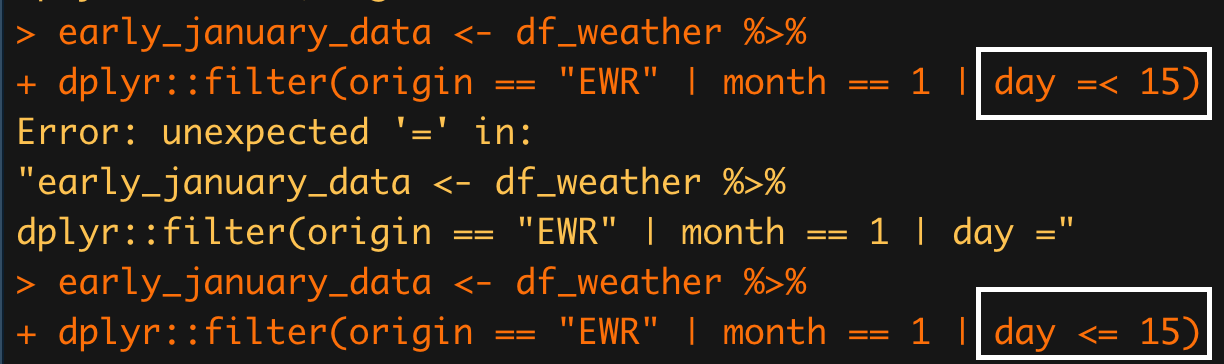
Figure C.6: less than or equal to
Response:
I suspect this is nothing more than a convention.
And the convention goes back several decades.
From wikipedia:
In BASIC, Lisp-family languages, and C-family languages (including Java and C++), operator <= means “less than or equal to.”
C.1.10 filter() and dplyr::filter()
I am trying to calculate the average of my new data set after excluding outliers using the
filterfunction.
I get the following message:
Error in df_sales_no_out$order_amount : $ operator is invalid for atomic vectorsAm I approaching this the wrong way?
Response:
filter() and dplyr::filter() are two different functions.
It appears that you wanted to use the second.
However, without referencing its package — dplyr,
R thinks that you are asking for the first one.
Here is the documentation
of filter() from the stats package.
You can see that it is far from what you’d want to use.
C.2 Tips
C.2.1 How to name objects in R
Recommended practice:
- all lower case letters.
Ris case sensitive.varis different fromVar. (How do you test?)
- use letter and numbers.
Special symbols are NOT allowed:$,@,!,^,+,-,/,*
Try for your self
- start with letter, not number
modle1is okay,1modleis not.
- choose meaningful names
df_flights, notdata, ordata1, ormydata
- connect words with underscore
C.2.2 Mis-spellings
I can recall numerous occasions when I spent hours trying to trouble shoot
some code,
only to find out in the end that I had mis-spelled some words.
R can quite unforgiving for mis-spellings.
There is no auto-correct.
If you mis-spell the name of an object or a function,
R will complain Error: object not found
or Error: could not find function.
- Words I have mis-spelled more than once in the past:
| mis-spelling | correct spelling | context |
|---|---|---|
| lables | labels | factor() |
| margrittr | magrittr | %>% |
| … |
C.2.3 renv
When updating renv.lock file using renv::snapshot(),
you may encounter messages like this:
WARNING: One or more problems were discovered while enumerating dependencies.
/xx/xx/xx.Rmd
-----------------------------------------------------------------------------------
ERROR 1: <text>:73:33: unexpected symbol
72: if (length(cran_primary) != 0) xf$pkg_load2(cran_primary)
73: if (length(cran_secondary != 0) xf
^
Please see `?renv::dependencies` for more information.
Do you want to proceed? [y/N]:You can safely ignore this error message and proceed with y.
Why are you seeing this message?
renvcrawls through all of your.Rmdand.Rfiles to discover what packages you are using and versions of those packages. Very smart, eh? However,renv’s smartness comes at a cost of being less flexible. It anticipates users to install packages and attach them withlibrary()and alike. However, as I mentioned in Section 1.3.3, I am not usinglibrary()and explicitly reference all functions with their respective packages.revndoes not expect this, or expressions likepackage$function, so will throw a warning whenever it finds such expressions.