Chapter 4 Data Importing and “Tidy” Data
In Subsection 1.2.1,
we introduced the concept of a data frame in R:
a rectangular spreadsheet-like representation of data
where the rows correspond to observations and the columns
correspond to variables describing each observation.
In Section 1.4, we started exploring our first data frame:
the flights data frame included in the nycflights13 package.
In Chapter 2, we created visualizations based on the data
included in flights and other data frames such as weather.
In Chapter 3, we learned how to take existing data frames
and transform/modify them to suit our ends.
In this final chapter of the “Data Science with tidyverse” portion
of the book,
we extend some of these ideas
by discussing a type of data formatting called “tidy” data.
You will see that having data stored in “tidy” format
is about more than just what the everyday definition
of the term “tidy” might suggest:
having your data “neatly organized.”
Instead, we define the term “tidy” as it’s used by data scientists who use R,
outlining a set of rules by which data is saved.
Knowledge of this type of data formatting was not necessary for our treatment of data visualization in Chapter 2 and data wrangling in Chapter 3. This is because all the data used were already in “tidy” format. In this chapter, we’ll now see that this format is essential to using the tools we covered up until now. Furthermore, it will also be useful for all subsequent chapters in this book when we cover regression and statistical inference. First, however, we’ll show you how to import spreadsheet data in R.
Needed packages
The following code gives us access to the demo data, as well as the to some tools for us to interact with the data.
# Install xfun so that I can use xfun::pkg_load2
if (!requireNamespace('xfun')) install.packages('xfun')
xf <- loadNamespace('xfun')
cran_primary <- c(
"dplyr",
"fivethirtyeight",
"ggplot2",
"here",
"nycflights13",
"readr",
"readxl",
"tidyr"
)
if (length(cran_primary) != 0) xf$pkg_load2(cran_primary)
import::from(magrittr, "%>%")
gg <- import::from(ggplot2, .all=TRUE, .into={new.env()})
dp <- import::from(dplyr, .all=TRUE, .into={new.env()})
import::from(nycflights13,
df_airlines = airlines,
df_airports = airports,
df_flights = flights,
df_planes = planes,
df_weather = weather
)4.1 Importing data
Up to this point, we’ve almost entirely used data stored inside of an R package. Say instead you have your own data saved on your computer or somewhere online. How can you analyze this data in R? Spreadsheet data is often saved in one of the following three formats:
First, a Comma Separated Values .csv file.
You can think of a .csv file as a bare-bones spreadsheet where:
- Each line in the file corresponds to one row of data/one observation.
- Values for each line are separated with commas. The values of different variables are separated by commas in each row.
- The first line is often, but not always, a header row indicating the names of the columns/variables.
Second, an Excel .xlsx spreadsheet file.
This format is based on Microsoft’s proprietary Excel software.
As opposed to bare-bones .csv files,
.xlsx Excel files contain a lot of meta-data
(data about data).
Recall we saw a previous example of meta-data in Section 3.5
when adding “group structure” meta-data to a data frame
by using the group_by() verb.
Some examples of Excel spreadsheet meta-data
include the use of bold and italic fonts, colored cells,
different column widths, and formula macros.
Third, a Google Sheets file,
which is a “cloud” or online-based way to work with a spreadsheet.
To import Google Sheets data in R,
you can use a package called
googlesheets,
a method we will leave for more advanced books.
Google Sheets also allows you to download your data
in both comma separated values .csv and Excel .xlsx formats.
Next we cover a method for importing .csv and .xlsx spreadsheet data in R.
4.1.1 .csv file
First, let’s import a Comma Separated Values .csv file
that exists on the internet.
The .csv file dem_score.csv contains ratings of the level of democracy
in different countries spanning 1952 to 1992 and
is accessible at https://moderndive.com/data/dem_score.csv.
Let’s use the read_csv() function from the readr
(Wickham and Hester 2020) package
to read it off the web, import it into R,
and save it in a data frame called dem_score.
# A tibble: 96 x 10
country `1952` `1957` `1962` `1967` `1972` `1977` `1982` `1987` `1992`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Albania -9 -9 -9 -9 -9 -9 -9 -9 5
2 Argentina -9 -1 -1 -9 -9 -9 -8 8 7
3 Armenia -9 -7 -7 -7 -7 -7 -7 -7 7
4 Australia 10 10 10 10 10 10 10 10 10
5 Austria 10 10 10 10 10 10 10 10 10
6 Azerbaijan -9 -7 -7 -7 -7 -7 -7 -7 1
7 Belarus -9 -7 -7 -7 -7 -7 -7 -7 7
8 Belgium 10 10 10 10 10 10 10 10 10
9 Bhutan -10 -10 -10 -10 -10 -10 -10 -10 -10
10 Bolivia -4 -3 -3 -4 -7 -7 8 9 9
# … with 86 more rowsIn this dem_score data frame,
the minimum value of -10 corresponds to a highly autocratic nation,
whereas a value of 10 corresponds to a highly democratic nation.
Note the backticks “`” that surround the different variable names.
Variable names in R by default are not allowed to start with a number
nor include spaces,
but for aesthetic purposes, it is occasionally desirable to have those “illegal”
variable names in a formated table.
We can achieve this by surrounding the “illegal” names with backticks.
We’ll revisit the dem_score data frame
in a case study in the upcoming Section 4.3.
Note that the read_csv() function included in the readr package
is different than the read.csv() function in base R,
which comes installed with R.
While the difference in the names might seem trivial (a _ instead of a .),
the read_csv() function is, in our opinion, easier to use
because it can more easily read data off the web
and generally imports data at a much faster speed.
Furthermore, the read_csv() function included in the readr
saves data frames as tibbles by default.
4.1.2 .xlsx file
Next, let’s import an Excel file, dem_score.xlsx.
This file is identical to dem_score.csv we just imported,
except that it has a different file extension.
To read in an excel file, we need to use read_excel() from packagereadxl.
Unlike read_csv() from readr,
read_excel() cannot directly read in a file from the internet.
Let’s download the file first.
If you are windows user:
data_url <- "https://moderndive.com/data/dem_score.xlsx"
download.file(url=data_url, destfile=here::here("data", "dem_score.xlsx"), mode="wb")If you are mac or linux user:
data_url <- "https://moderndive.com/data/dem_score.xlsx"
xfun::download_file(url=data_url, output = here::here("data", "dem_score.xlsx"))Now dem_score.xlsx should be present in the “data” foler inside your project.
Let’s import this data into R now.
4.2 “Tidy” data
Let’s now switch gears and learn about the concept of “tidy” data format
with a motivating example from the fivethirtyeight package.
The fivethirtyeight package (Kim, Ismay, and Chunn 2020) provides access
to the datasets used in many articles published
by the data journalism website,
FiveThirtyEight.com.
For a complete list of all 129
datasets included in the fivethirtyeight package,
check out the package webpage by going to:
https://fivethirtyeight-r.netlify.app/articles/fivethirtyeight.html.
Let’s focus our attention on the drinks data frame
and look at its first 5 rows:
# A tibble: 6 x 5
country beer_servings spirit_servings wine_servings total_litres_of_pure…
<chr> <int> <int> <int> <dbl>
1 Afghanistan 0 0 0 0
2 Albania 89 132 54 4.9
3 Algeria 25 0 14 0.7
4 Andorra 245 138 312 12.4
5 Angola 217 57 45 5.9
6 Antigua & B… 102 128 45 4.9After reading the help file by running ?fivethirtyeight::drinks,
you’ll see that df_drinks is a data frame
containing results from a survey of the average number of servings of beer,
spirits, and wine consumed in 193 countries.
This data was originally reported on FiveThirtyEight.com
in Mona Chalabi’s article:
“Dear Mona Followup: Where Do People Drink The Most Beer, Wine And Spirits?”.
Let’s apply some of the data wrangling verbs we learned
in Chapter 3 on the df_drinks data frame:
filter()thedf_drinksdata frame to only consider 4 countries: the United States, China, Italy, and Saudi Arabia, thenselect()all columns excepttotal_litres_of_pure_alcoholby using the-sign, thenrename()the variablesbeer_servings,spirit_servings, andwine_servingstobeer,spirit, andwine, respectively.
and save the resulting data frame in drinks_smaller:
drinks_smaller <- df_drinks %>%
dp$filter(country %in% c("USA", "China", "Italy", "Saudi Arabia")) %>%
dp$select(-total_litres_of_pure_alcohol) %>%
dp$rename(beer = beer_servings, spirit = spirit_servings, wine = wine_servings)
drinks_smaller# A tibble: 4 x 4
country beer spirit wine
<chr> <int> <int> <int>
1 China 79 192 8
2 Italy 85 42 237
3 Saudi Arabia 0 5 0
4 USA 249 158 84Let’s now ask ourselves a question:
“Using the drinks_smaller data frame,
how would we create the side-by-side barplot
in Figure 4.1?”
Recall we saw barplots displaying two categorical variables
in Subsection 2.8.3.
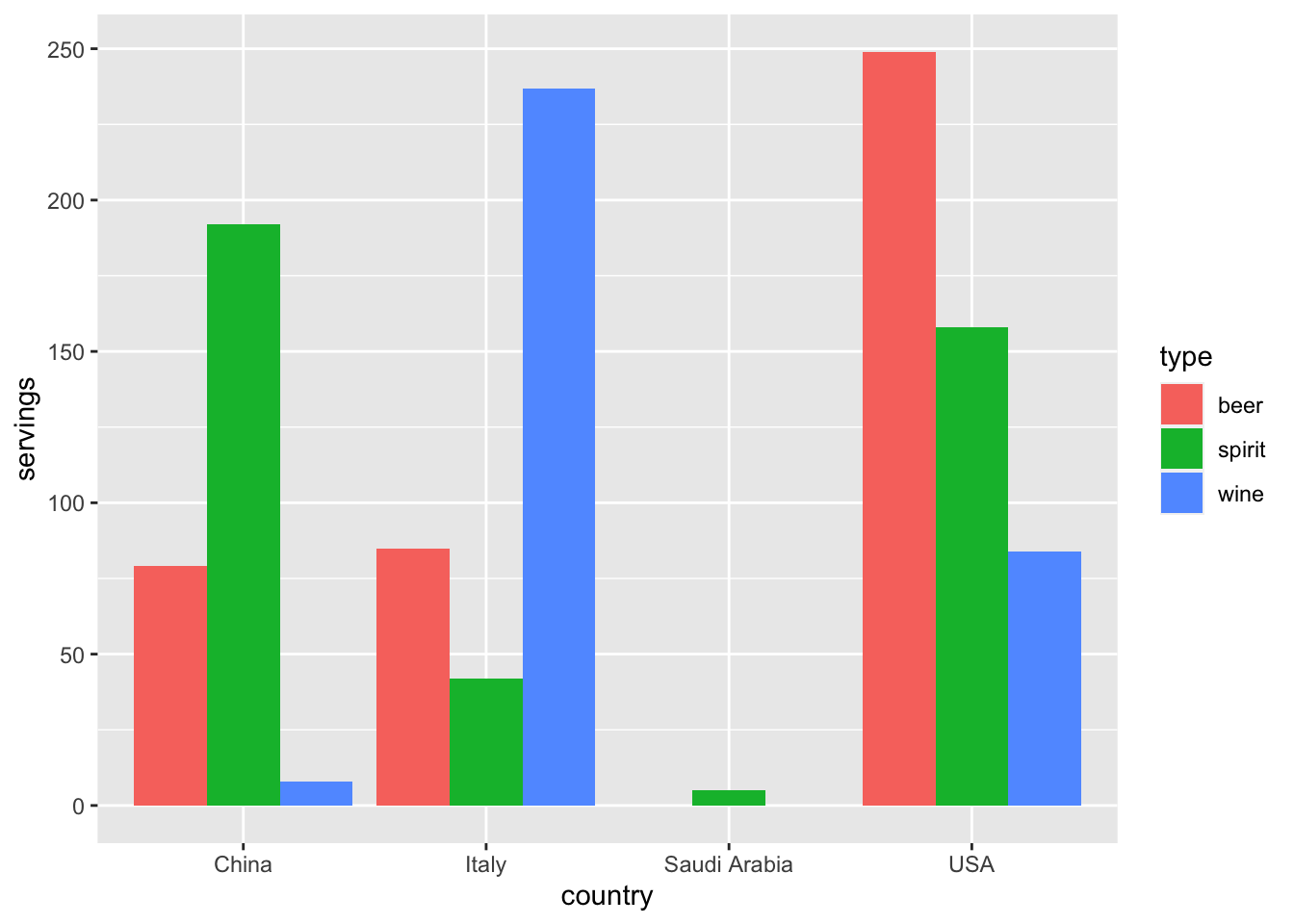
Figure 4.1: Comparing alcohol consumption in 4 countries.
Let’s break down the grammar of graphics we introduced in Section 2.1:
- The categorical variable
countrywith four levels (China, Italy, Saudi Arabia, USA) would have to be mapped to thex-position of the bars. - The numerical variable
servingswould have to be mapped to they-position of the bars (the height of the bars). - The categorical variable
typewith three levels (beer, spirit, wine) would have to be mapped to thefillcolor of the bars.
Observe that drinks_smaller has three separate variables
beer, spirit, and wine.
In order to use the ggplot() function to recreate the barplot
in Figure 4.1 however,
we need a single variable type with three possible values:
beer, spirit, and wine.
We could then map this type variable to the fill aesthetic of our plot.
In other words, to recreate the barplot in Figure 4.1,
our data frame would have to look like this:
# A tibble: 12 x 3
country type servings
<chr> <chr> <int>
1 China beer 79
2 China spirit 192
3 China wine 8
4 Italy beer 85
5 Italy spirit 42
6 Italy wine 237
7 Saudi Arabia beer 0
8 Saudi Arabia spirit 5
9 Saudi Arabia wine 0
10 USA beer 249
11 USA spirit 158
12 USA wine 84Observe that while drinks_smaller and drinks_smaller_tidy
are both rectangular in shape
and contain the same 12 numerical values
(3 alcohol types by 4 countries),
they are formatted differently.
drinks_smaller is formatted in what’s known as
“wide” format,
whereas drinks_smaller_tidy is formatted in what’s known as
“long/narrow” format.
In the context of doing data science in R,
long/narrow format is also known as “tidy” format.
In order to use the ggplot2 and dplyr packages
for data visualization and data wrangling,
your input data frames must be in “tidy” format.
Thus, all non-“tidy” data must be converted to “tidy” format first.
Before we convert non-“tidy” data frames
like drinks_smaller to “tidy” data frames like drinks_smaller_tidy,
let’s define “tidy” data.
4.2.1 Definition of “tidy” data
You have surely heard the word “tidy” in your life:
- “Tidy up your room!”
- “Write your homework in a tidy way so it is easier to provide feedback.”
- Marie Kondo’s best-selling book, The Life-Changing Magic of Tidying Up: The Japanese Art of Decluttering and Organizing, and Netflix TV series Tidying Up with Marie Kondo.
- “I am not by any stretch of the imagination a tidy person, and the piles of unread books on the coffee table and by my bed have a plaintive, pleading quality to me - ‘Read me, please!’” - Linda Grant
What does it mean for your data to be “tidy?” While “tidy” has a clear English meaning of “organized,” the word “tidy” in data science using R means that your data follows a standardized format. We will follow Hadley Wickham’s definition of “tidy” data (Wickham 2014) shown also in Figure 4.2:

Figure 4.2: Tidy data graphic from R for Data Science.
A dataset is a collection of values, usually either numbers (if quantitative) or strings AKA text data (if qualitative/categorical). Values are organised in two ways. Every value belongs to a variable and an observation. A variable contains all values that measure the same underlying attribute (like height, temperature, duration) across units. An observation contains all values measured on the same unit (like a person, or a day, or a city) across attributes.
“Tidy” data is a standard way of mapping the meaning of a dataset to its structure. A dataset is messy or tidy depending on how rows, columns and tables are matched up with observations, variables and types. In tidy data:
- Each variable forms a column.
- Each observation forms a row.
- Each type of observational unit forms a table.
For example, say you have the following table of stock prices in Table 4.1:
| Date | Boeing stock price | Amazon stock price | Google stock price |
|---|---|---|---|
| 2009-01-01 | $173.55 | $174.90 | $174.34 |
| 2009-01-02 | $172.61 | $171.42 | $170.04 |
Although the data are neatly organized in a rectangular spreadsheet-type format, they do not follow the definition of data in “tidy” format. Although there are three variables corresponding to three unique pieces of information (date, stock name, and stock price), the three variables are not organized in three columns. In “tidy” data format, each variable should be its own column, as shown in Table 4.2. Notice that both tables present the same information, but in different formats.
| Date | Stock Name | Stock Price |
|---|---|---|
| 2009-01-01 | Boeing | $173.55 |
| 2009-01-01 | Amazon | $174.90 |
| 2009-01-01 | $174.34 | |
| 2009-01-02 | Boeing | $172.61 |
| 2009-01-02 | Amazon | $171.42 |
| 2009-01-02 | $170.04 |
Now we have the requisite three columns Date, Stock Name, and Stock Price. On the other hand, consider the data in Table 4.3.
| Date | Boeing Price | Weather |
|---|---|---|
| 2009-01-01 | $173.55 | Sunny |
| 2009-01-02 | $172.61 | Overcast |
In this case, even though the variable “Boeing Price” occurs just like in our non-“tidy” data in Table 4.1, the data is “tidy” because there are three variables corresponding to three unique pieces of information: Date, Boeing price, and the Weather on that particular day.
Learning check
(LC4.1) What are common characteristics of “tidy” data frames?
4.2.2 Converting to “tidy” data
In this book so far, you’ve only seen data frames
that were already in “tidy” format.
Furthermore, for the rest of this book,
you’ll mostly only see data frames that are already in “tidy” format as well.
This is not always the case however with all datasets in the world.
If your original data frame is in wide (non-“tidy”) format
and you would like to use the ggplot2 or dplyr packages,
you will first have to convert it to “tidy” format.
To do so, we recommend using the pivot_longer()
function in the tidyr package (Wickham 2020).
Going back to our drinks_smaller data frame from earlier:
# A tibble: 4 x 4
country beer spirit wine
<chr> <int> <int> <int>
1 China 79 192 8
2 Italy 85 42 237
3 Saudi Arabia 0 5 0
4 USA 249 158 84We can convert it to “tidy” format by using the pivot_longer() function
from the tidyr package as follows:
drinks_smaller_tidy <- drinks_smaller %>%
tidyr::pivot_longer(names_to = "type",
values_to = "servings",
cols = -country)
drinks_smaller_tidy# A tibble: 12 x 3
country type servings
<chr> <chr> <int>
1 China beer 79
2 China spirit 192
3 China wine 8
4 Italy beer 85
5 Italy spirit 42
6 Italy wine 237
7 Saudi Arabia beer 0
8 Saudi Arabia spirit 5
9 Saudi Arabia wine 0
10 USA beer 249
11 USA spirit 158
12 USA wine 84We set the arguments to pivot_longer() as follows:
names_tohere corresponds to the name of the variable in the new “tidy”/long data frame that will contain the column names of the original data. Observe how we setnames_to = "type". In the resultingdrinks_smaller_tidy, the columntypecontains the three types of alcoholbeer,spirit, andwine. Sincetypeis a variable name that doesn’t appear indrinks_smaller, we use quotation marks around it. You’ll receive an error if you just usenames_to = typehere.values_tohere is the name of the variable in the new “tidy” data frame that will contain the values of the original data. Observe how we setvalues_to = "servings"since each of the numeric values in each of thebeer,wine, andspiritcolumns of thedrinks_smallerdata corresponds to a value ofservings. In the resultingdrinks_smaller_tidy, the columnservingscontains the 4 \(\times\) 3 = 12 numerical values. Note again thatservingsdoesn’t appear as a variable indrinks_smallerso it again needs quotation marks around it for thevalues_toargument.- The third argument
colsis the columns in thedrinks_smallerdata frame you either want to or don’t want to “tidy.” Observe how we set this to-countryindicating that we don’t want to “tidy” thecountryvariable indrinks_smaller. As a result, onlybeer,spirit, andwinecolumns from the original data frame will be transformed. Becausecountryis an existing column from the original data framedrinks_smaller, we don’t put quotation marks around it.
The argument cols can be specified in more than one way.
Let’s consider code that’s written slightly differently
but that produces the same output:
drinks_smaller %>%
tidyr::pivot_longer(names_to = "type",
values_to = "servings",
cols = c(beer, spirit, wine))Note that the third argument now specifies which columns we want to “tidy”:
c(beer, spirit, wine),
instead of the columns we don’t want to “tidy”: -country.
We use the c() function to create a vector of the columns in drinks_smaller
that we’d like to “tidy.”
Note that because these three columns are adjacent to each other,
in the drinks_smaller data frame,
we could also do the following for the cols argument:
With our drinks_smaller_tidy “tidy” formatted data frame, we can now produce the barplot you saw in Figure 4.1 using geom_col(). This is done in Figure 4.3. Recall from Section 2.8 on barplots that we use geom_col() and not geom_bar(), since we would like to map the “pre-counted” servings variable to the y-aesthetic of the bars.
gg$ggplot(drinks_smaller_tidy, gg$aes(x = country, y = servings, fill = type)) +
gg$geom_col(position = "dodge")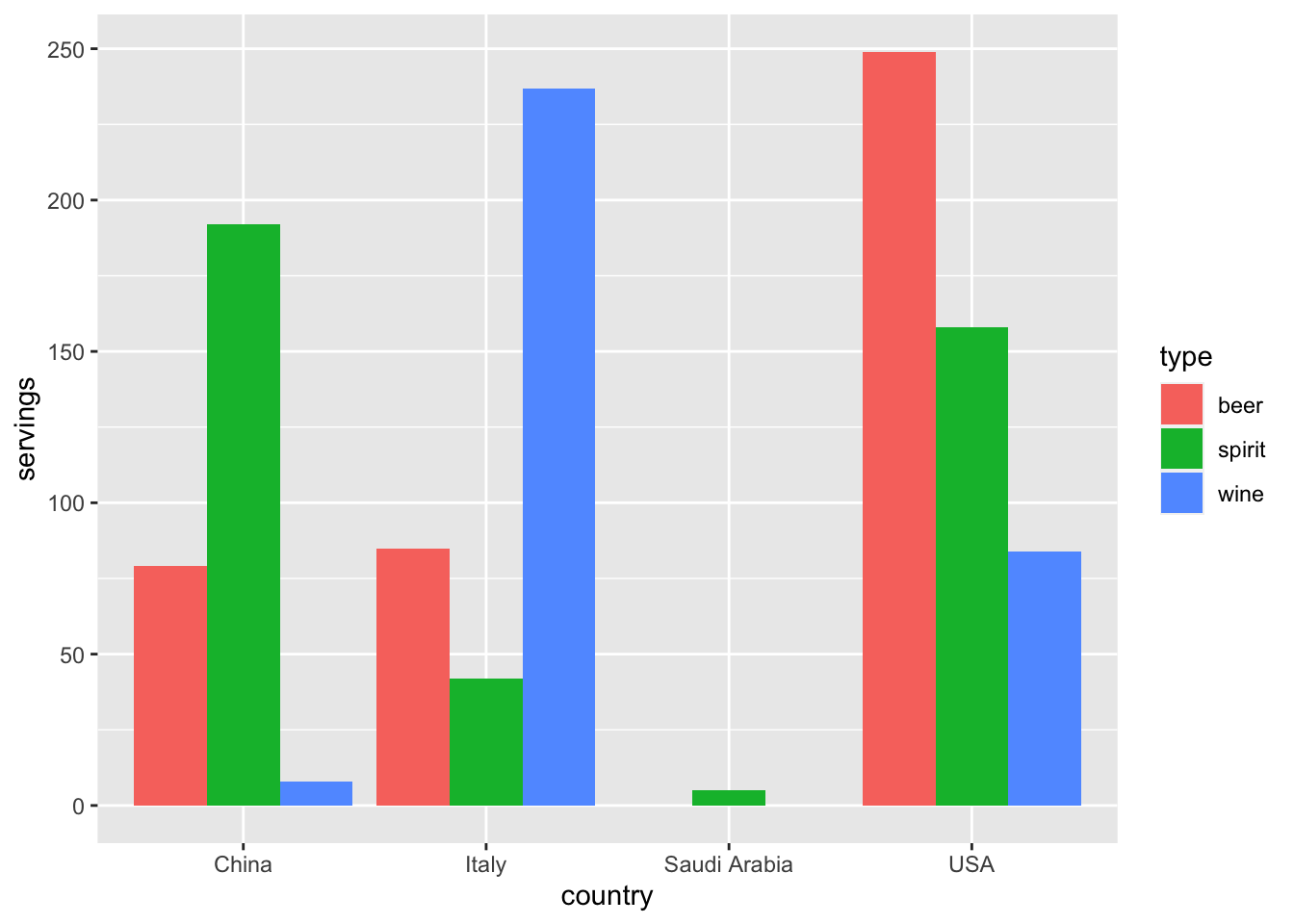
Figure 4.3: Comparing alcohol consumption in 4 countries using geom_col().
Converting “wide” format data to “tidy” format often confuses new R users.
The only way to learn to get comfortable with the pivot_longer() function
is with practice, practice, and more practice using different datasets.
For example, run ?tidyr::pivot_longer
and look at the examples in the bottom of the help file.
We’ll show another example of using pivot_longer()
to convert a “wide” formatted data frame to “tidy” format
in Section 4.3.
If however you want to convert a “tidy” data frame to “wide” format,
you will need to use the pivot_wider() function instead.
Run ?tidyr::pivot_wider and look at the examples
in the bottom of the help file for examples.
You can also view examples of both pivot_longer() and pivot_wider()
on the tidyverse.org webpage.
There’s a nice example to check out the different functions available for data tidying and a case study using data from the World Health Organization on that webpage. Furthermore, each week the R4DS Online Learning Community posts a dataset in the weekly #TidyTuesday event that might serve as a nice place for you to find other data to explore and transform.
Learning check
(LC4.3)
Take a look at the airline_safety data frame
included in the fivethirtyeight data package.
Run the following:
# A tibble: 56 x 9
airline incl_reg_subsid… avail_seat_km_p… incidents_85_99 fatal_accidents…
<chr> <lgl> <dbl> <int> <int>
1 Aer Li… FALSE 320906734 2 0
2 Aerofl… TRUE 1197672318 76 14
3 Aeroli… FALSE 385803648 6 0
4 Aerome… TRUE 596871813 3 1
5 Air Ca… FALSE 1865253802 2 0
6 Air Fr… FALSE 3004002661 14 4
7 Air In… TRUE 869253552 2 1
8 Air Ne… TRUE 710174817 3 0
9 Alaska… TRUE 965346773 5 0
10 Alital… FALSE 698012498 7 2
# … with 46 more rows, and 4 more variables: fatalities_85_99 <int>,
# incidents_00_14 <int>, fatal_accidents_00_14 <int>, fatalities_00_14 <int>After reading the help file by running ?fivethirtyeight::airline_safety,
we see that airline_safety is a data frame
containing information on different airline companies’ safety records.
This data was originally reported on the data journalism website,
FiveThirtyEight.com, in Nate Silver’s article,
“Should Travelers Avoid Flying Airlines That Have Had Crashes in the Past?”.
Let’s only consider the variables airlines
and those relating to fatalities for simplicity:
airline_safety_smaller <- df_airline_safety %>%
dp$select(airline, dp$starts_with("fatalities"))
airline_safety_smaller# A tibble: 56 x 3
airline fatalities_85_99 fatalities_00_14
<chr> <int> <int>
1 Aer Lingus 0 0
2 Aeroflot 128 88
3 Aerolineas Argentinas 0 0
4 Aeromexico 64 0
5 Air Canada 0 0
6 Air France 79 337
7 Air India 329 158
8 Air New Zealand 0 7
9 Alaska Airlines 0 88
10 Alitalia 50 0
# … with 46 more rowsThis data frame is not in “tidy” format. How would you convert this data frame to be in “tidy” format, in particular so that it has a variable fatalities_years indicating the incident year and a variable num of the fatality counts?
4.2.3 Data frames in nycflights13
Recall the nycflights13 package
we introduced in Section 1.4
with data about all domestic flights departing from New York City in 2013.
Let’s revisit the df_flights data frame by running View(df_flights).
We saw that df_flights has a rectangular shape,
with each of its 336,776
rows corresponding to a flight and each of its 19
columns corresponding to different characteristics/measurements of each flight.
This satisfied the first two criteria of the definition
of “tidy” data from Subsection 4.2.1:
that “Each variable forms a column” and “Each observation forms a row.”
But what about the third property of “tidy” data
that “Each type of observational unit forms a table?”
Recall that we saw in Subsection 1.4.2
that the observational unit for the df_flights data frame
is an individual flight.
In other words, the rows of the df_flights data frame refer to characteristics/measurements of individual flights. Also included in the nycflights13 package are other data frames with their rows representing different observational units (Wickham 2019):
airlines: translation between two letter IATA carrier codes and airline company names (16 in total). The observational unit is an airline company.planes: aircraft information about each of 3,322 planes used, i.e., the observational unit is an aircraft.weather: hourly meteorological data (about 8,705 observations) for each of the three NYC airports, i.e., the observational unit is an hourly measurement of weather at one of the three airports.airports: airport names and locations. The observational unit is an airport.
The organization of the information into these five data frames follows the third “tidy” data property: observations corresponding to the same observational unit should be saved in the same table, i.e., data frame. You could think of this property as the old English expression: “birds of a feather flock together.”
4.3 Case study: Democracy in Guatemala
In this section, we’ll show you another example of how to convert a data frame that isn’t in “tidy” format (“wide” format) to a data frame that is in “tidy” format (“long/narrow” format). We’ll do this using the pivot_longer() function from the tidyr package again.
Furthermore, we’ll make use of functions from the ggplot2 and dplyr packages to produce a time-series plot showing how the democracy scores have changed over the 40 years from 1952 to 1992 for Guatemala. Recall that we saw time-series plots in Section 2.4 on creating linegraphs using geom_line().
Let’s use the dem_score data frame we imported in Section 4.1, but focus on only data corresponding to Guatemala.
# A tibble: 1 x 10
country `1952` `1957` `1962` `1967` `1972` `1977` `1982` `1987` `1992`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Guatemala 2 -6 -5 3 1 -3 -7 3 3Let’s lay out the grammar of graphics we saw in Section 2.1.
First we know we need to set data = guat_dem and use a geom_line() layer, but what is the aesthetic mapping of variables? We’d like to see how the democracy score has changed over the years, so we need to map:
yearto the x-position aesthetic anddemocracy_scoreto the y-position aesthetic
Now we are stuck in a predicament, much like with our drinks_smaller example in Section 4.2. We see that we have a variable named country, but its only value is "Guatemala". We have other variables denoted by different year values. Unfortunately, the guat_dem data frame is not “tidy” and hence is not in the appropriate format to apply the grammar of graphics, and thus we cannot use the ggplot2 package just yet.
We need to take the values of the columns corresponding to years in guat_dem and convert them into a new “names” variable called year. Furthermore, we need to take the democracy score values in the inside of the data frame and turn them into a new “values” variable called democracy_score. Our resulting data frame will have three columns: country, year, and democracy_score. Recall that the pivot_longer() function in the tidyr package does this for us:
guat_dem_tidy <- guat_dem %>%
tidyr::pivot_longer(names_to = "year",
values_to = "democracy_score",
cols = -country,
names_transform = list(year = as.integer))
guat_dem_tidy# A tibble: 9 x 3
country year democracy_score
<chr> <int> <dbl>
1 Guatemala 1952 2
2 Guatemala 1957 -6
3 Guatemala 1962 -5
4 Guatemala 1967 3
5 Guatemala 1972 1
6 Guatemala 1977 -3
7 Guatemala 1982 -7
8 Guatemala 1987 3
9 Guatemala 1992 3We set the arguments to pivot_longer() as follows:
names_tois the name of the variable in the new “tidy” data frame that will contain the column names of the original data. Observe how we setnames_to = "year". In the resultingguat_dem_tidy, the columnyearcontains the years where Guatemala’s democracy scores were measured.values_tois the name of the variable in the new “tidy” data frame that will contain the values of the original data. Observe how we setvalues_to = "democracy_score". In the resultingguat_dem_tidythe columndemocracy_scorecontains the 1 \(\times\) 9 = 9 democracy scores as numeric values.- The third argument is the columns you either want to or don’t want to “tidy.” Observe how we set this to
cols = -countryindicating that we don’t want to “tidy” thecountryvariable inguat_demand rather only variables1952through1992. - The last argument of
names_transformtells R what type of variableyearshould be set to. Without specifying that it is anintegeras we’ve done here,pivot_longer()will set it to be a character value by default.
We can now create the time-series plot in Figure 4.4 to visualize how democracy scores in Guatemala have changed from 1952 to 1992 using a geom_line(). Furthermore, we’ll use the labs() function in the ggplot2 package to add informative labels to all the aes()thetic attributes of our plot, in this case the x and y positions.
gg$ggplot(guat_dem_tidy, gg$aes(x = year, y = democracy_score)) +
gg$geom_line() +
gg$labs(x = "Year", y = "Democracy Score")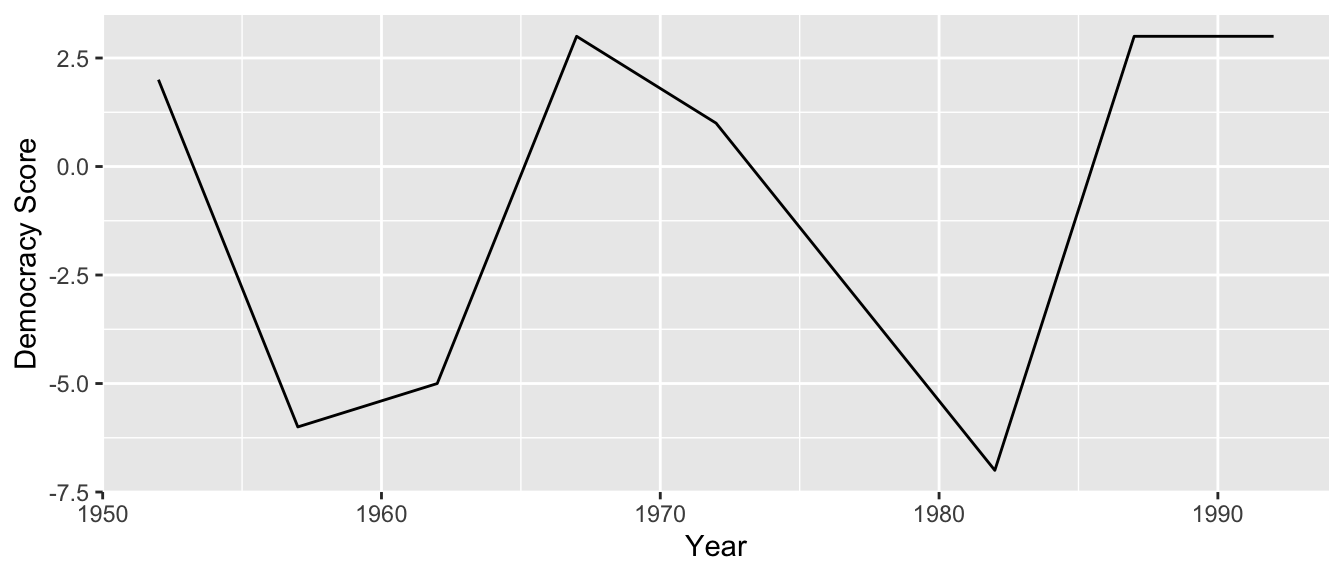
Figure 4.4: Democracy scores in Guatemala 1952-1992.
Note that if we forgot to include the names_transform argument specifying that year was not of character format, we would have gotten an error here since geom_line() wouldn’t have known how to sort the character values in year in the right order.
Learning check
(LC4.4) Convert the dem_score data frame into
a “tidy” data frame and assign the name of dem_score_tidy to the resulting long-formatted data frame.
(LC4.5)
Read in the life expectancy data stored at
https://moderndive.com/data/le_mess.csv
and convert it to a “tidy” data frame.
Hint: use readr::read_csv() function we learned earlier
to import the data.
4.4 tidyverse package
So far I have managed to avoid discussing the elephant in the room:
tidyverse.
However, you must have seen it in Datacamp and many other references.
What is tidyverse?
It is an amalgamation of several packages,
each specilizing on one aspect of data manipulation.
So far, we have seen four of them in this course:
ggplot2 for data visualization,
dplyr for data wrangling,
readr for importing spreadsheet data into R,
and tidyr for converting data to “tidy” format.
The tidyverse “umbrella” package
gets its name from the fact that all the functions in all its packages are designed to have common inputs and outputs: data frames are in “tidy” format. This standardization of input and output data frames makes transitions between different functions in the different packages as seamless as possible.
tidyverse was created mainly for convience purposes.
Instead of having to install many different packages,
one can install tidyverse and have access to all of them.
However, I have chosen to stick with individual packages for this course.
This choice was partly inspired by my own experience:
it is easier to remember what each function does
once I have learned their grouping under their respective packages
(remember “chunking” from your psyc101?).
Sooner or later, you will encounter R scripts
that start with install.packages("tidyverse"),
or library(tidyverse).
I recommend that you avoid using this umbrella package in your own code
and always look functions up whenever you are unsure which packages they belongs to.
Do so until you can comfortably identify the membership
of most commonly used functions
and are used to resorting to help whenever you are unsure.
4.5 Conclusion
4.5.1 Additional resources
4.5.2 What’s to come?
Congratulations! You’ve completed the “Data Science with tidyverse” portion of this book. We’ll now move to the “Data modeling with moderndive” portion of this book in Chapters 11 and 14, where you’ll leverage your data visualization and wrangling skills to model relationships between different variables in data frames.
However, we’re going to leave Chapter 12 on “Inference for Regression” until after we’ve covered statistical inference in Chapters 5, 6, and 7. Onwards and upwards into Data Modeling as shown in Figure 4.5!
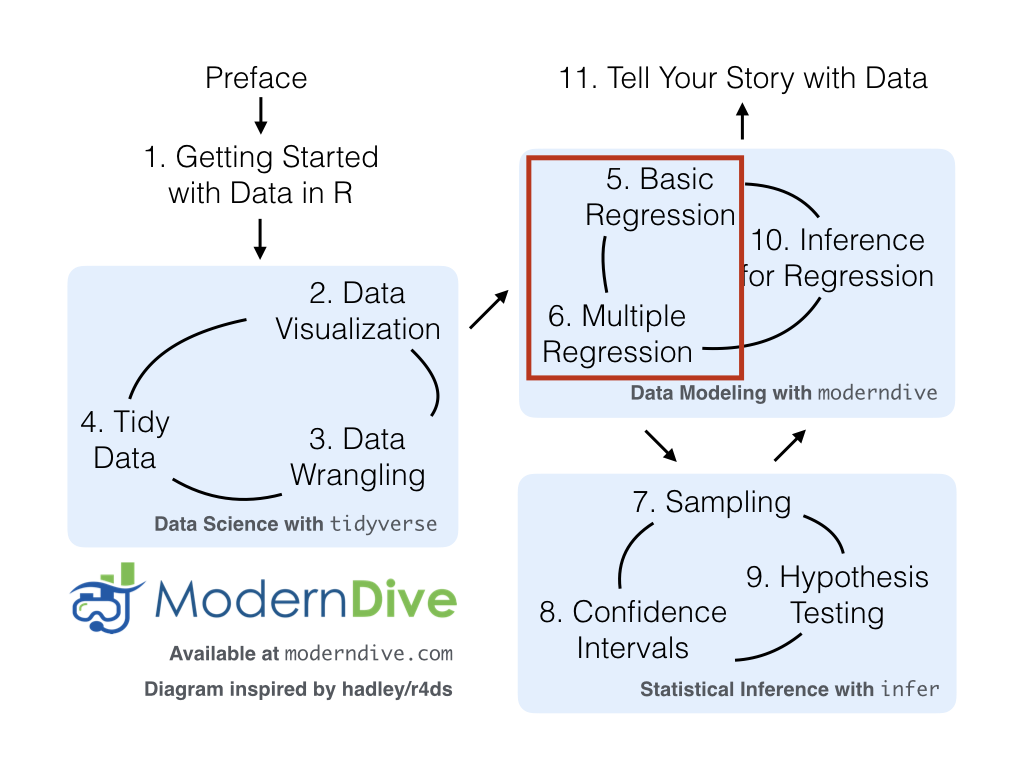
Figure 4.5: ModernDive flowchart - on to Part II!